Biology Reference
In-Depth Information
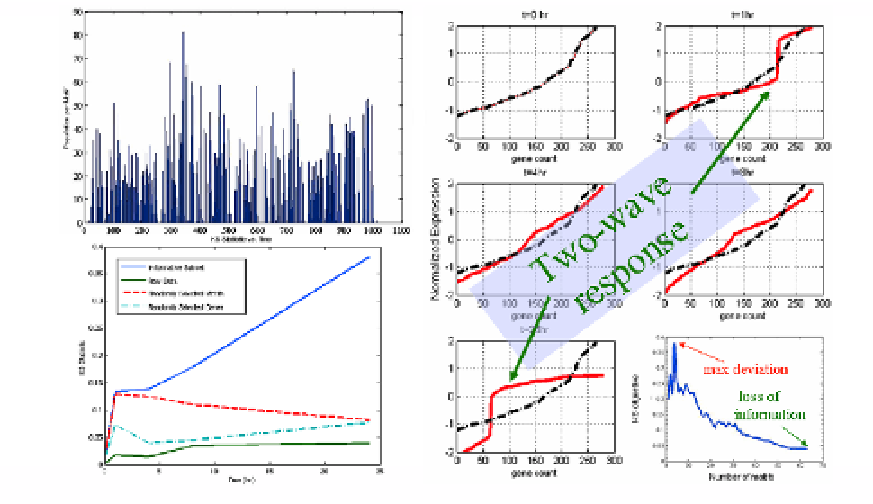
Fig. 3.2.
Basic elements of the process for selecting informative expression profiles.
equiprobable discretization is applied where the breakpoints are defined such that
the area defined by the boundaries of the breakpoint are equal. This method of
discretization was selected because empirical evidence suggests that the z-score
normalized sub-patterns should have a highly Gaussian distribution [46], thereby
equally distributing a set of randomly generated signals throughout the hash space.
Coefficients below the smallest breakpoint are “mapped” to the first symbol of a
chosen alphabet. Other points are “mapped” accordingly within their respective
intervals. A more extensive discussion and visualization of this process and can
be found in [46].
(b) Identification of informative expression motifs. This novel symbolic rep-
resentation makes it possible to further simplify the time series in order to
uniquely characterize the overall dynamic response of each transcriptional pro-
file with a single identifier [47]. After the alphabet has been generated, it is con-
densed into a single value using the function proposed by [48]:
hash
(
c,w,a
)=
1+Σ
j
=1
[
ord
(
c
j
)
a
w−j
, where a is the size of the alphabet, w is length of
the word, and c is the “letter” sequence to which the expression profile is assigned.
The parameter a is selected such that the population distribution of the motifs ex-
hibits significant non-exponential distribution signaling the presence of significant
differences in the population of expression profiles. Genes with similar normal-
ized expression profiles “hash” to similar motif values to generate a distribution
−
1]
∗









Search WWH ::

Custom Search