Biology Reference
In-Depth Information
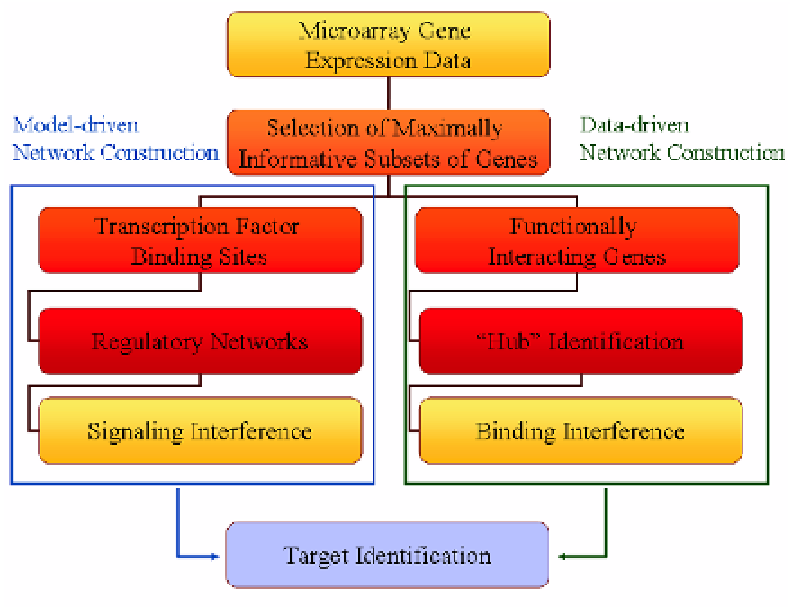
Fig. 3.1.
Functional and regulatory networks.
3.1.1.
Identification of Informative Temporal Expression Patterns
In order to develop context-specific networks, that is networks characteristic of
cellular responses to specific external stimuli, we need to first identify a sub-
set of relevant co-expressed genes whose transcription profiles are maximally af-
fected by the specific experiment performed. We recently proposed an integrated
methodology is composed of a number of steps outlined in Fig. 3.2, [45] for the
analysis of temporal gene expression measurements.
Specifically, our analysis
consists of the following steps:
(a) Fine-grained clustering. Each gene's expression is characterized as a wave-
form over time and a characteristic attribute is defined for each time course. This
identifier is then used to compare waveforms to each other and group similar
waveforms, based on the similarity of the identifiers, to characteristic expression
motifs. We have adopted the basic formalism of Symbolic Aggregate approX-
imation of the time series discussed in [46]. SAX is based on the premise of
transforming a time series into a corresponding sequence of symbols. Each series
is first normalized via the z-score given as
Y
j,t
=
Y
j,t
−µ
σ
with j=genes, t=time. An










Search WWH ::

Custom Search