Biology Reference
In-Depth Information
combined since the dissimilarity of these two clusters are the minimum among all
mutual dissimilarities of remaining clusters. This procedure stops when there is
only one cluster left, i.e., all objects are clustered into one cluster.
There are several variants of the agglomerative hierarchical clustering algo-
rithm. The selection of dissimilarity measure between two clusters is one source
of the variants. Another source of the variants is the stopping criteria. Well-known
variants of hierarchical clustering methods include CURE [11], ROCK [12], and
CHEMELEON [18].
The dissimilarity measure of two clusters used in the agglomerative hierarchi-
cal clustering algorithm includes single-linkage dissimilarity, complete-linkage
dissimilarity, minimum-variance dissimilarity [30], and some others [15]. The
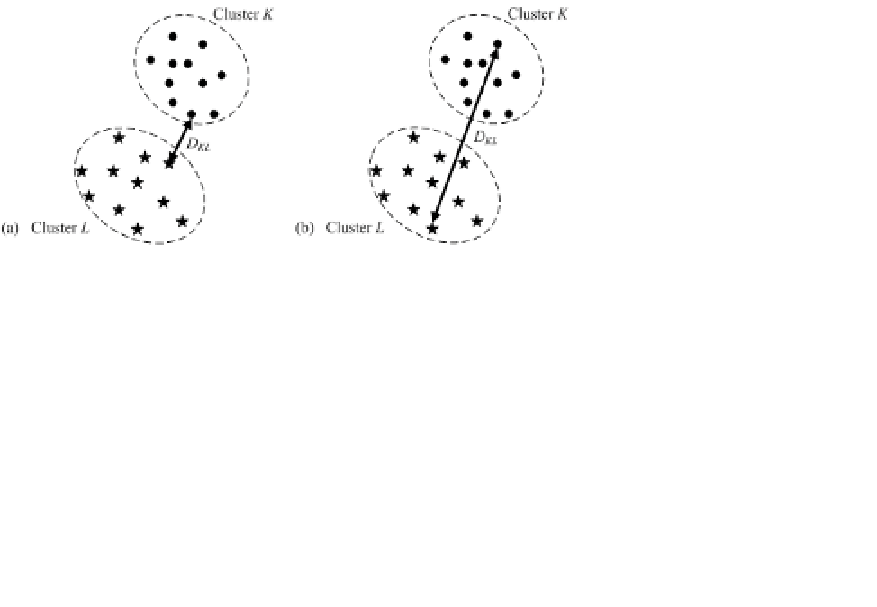
first two are most popularly used measures. Figure 5.4 illustrates these two mea-
sures with a 2-D example. Single-linkage dissimilarity of two clusters is the min-
imum of the distances between all pairs of objects, one from a cluster, and the
other one from another cluster. In Fig. 5.4, we use
D
KL
to represent the dissimilar-
ity between clusters
K
and
L
, denoted as
C
K
and
C
L
respectively. Single-linkage
dissimilarity is calculated as follows:
D
KL
=min
x
i
∈
C
K
,
x
j
∈
C
L
d
2
(
x
i
,
x
j
);
∀
i
,
j
.
(5.17)
Contrarily, complete-linkage dissimilarity between clusters
C
K
and
C
L
takes
the maximum one as the dissimilarity measure, i.e.,
D
KL
=m x
x
i
∈
C
K
,
x
j
∈
C
L
d
2
(
x
i
,
x
j
);
∀
i
,
j
.
(5.18)
Fig. 5.4.
Dissimilarity measure of two clusters: (a)single-linkage; (b)complete linkage.
Now, we compare the performance of these two dissimilarity measures in hier-
archical clustering algorithms to help users to determine which measure to choose
when clustering different datasets. Jain and Goel [15] compare the performance
of these two dissimilarity measures and several other measures in agglomerative







Search WWH ::

Custom Search