Biology Reference
In-Depth Information
u
.
.
.
ACGTTGAACTGCT
TGACGTTGAACTG
.
.
.
.
.
Part V
i
w
uv
v
Fig. 4.1. Graph representation for the motif finding problem. A graph part
V
i
corresponds to every
sequence, and a vertex corresponds to every possible motif position. If the motif length
l
is four, the
picture matches up the last few substrings to their graph vertices. Every pair of vertices
u
and
v
in
distinct parts are connected by an edge
w
uv
.
and Sze [35]. In this formulation an unknown pattern of a given length is inserted
with modifications into each of the input sequences. The positions of insertion
are unknown, as are the modifications in the instances of the pattern. Pevzner
and Sze focus on what they call the (
l,d
)-signal version, in which the pattern is a
string of length
l
and each instance differs from the pattern in exactly
d
positions.
The mutations are allowed to occur anywhere in the pattern, and thus any two
instances of the pattern may differ from one another in as many as 2
d
positions.
On the other hand, if two substrings differ in more than 2
d
positions, they cannot
simultaneously be instances of the implanted pattern. The graph version of the
problem remains largely the same as above, except that it is no longer a complete
N
-partite graph. By definition, vertices that correspond to substrings separated by
a Hamming distance that is greater than 2
d
, are not connected by an edge, as such
a vertex pair can not be an instantiation of a single pattern. The weights on the
edges are distance rather than similarity-based and are computed by considering
the number of matches and mismatches between the substrings.
We first address the algorithms designed for the standard motif finding prob-
lem, and then review some of the subtle motif methods.











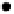






Search WWH ::

Custom Search