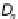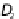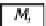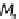Information Technology Reference
In-Depth Information
mean and the standard deviation of all the values for the feature
x
i
.Inthiswork
we only focused on data normalization but much more can be done to improve the
quality of the models [2].
Learning Ensemble Models.
One of the main advantages of ensemble learn-
ing methods is that they permit achieving predictions of better quality than those
obtained by standalone models. For generating the models we use the Bootstrap
Aggregating (Bagging) technique [17]. This technique reduces the
variance
, i.e.
the expected error derived from all the possible training sets for the problem.
The Bagging technique works as follows. For a given training dataset
D
,
n
D
i
) of size
m
are obtained by sampling the set
new training datasets (
ran-
domly with replacement (some examples are removed and some are repeated).
Each of the
n
bootstrap samples are used to generate
n
different (base) models.
The outputs of the
n
models are combined by averaging them. This procedure
generates a combined model that usually outperforms the single models and is
never considerably worse. As base models we use M5P regression trees, which
were discussed in the previous paragraphs. The entire process is illustrated in
Figure 3a.
D
(a)
Bagging process. The
n
bootstrap samples
(
D
i
) are used to construct the base models (
M
i
).
The base models are M5P regression trees.
(b)
Example of an M5P tree con-
structed for one of the
D
i
bootstrap
samples.
Fig. 3.
Process for Bagging M5P regression trees
4 Experiment Settings
To analyze the performance of the ensemble models we evaluated the predictive
accuracy of standalone models generated using the reviewed methods and en-
semble models learned with the Bagging strategy. Following sections describe a
bioinformatics application used as case of study, and details the methodology
used for the validation of our proposal.
4.1 Gene Expression Analysis Workflows
For the purposes of this work we evaluated our approach on bioinformatics data-
mining workflows, which perform a large-scale gene expression analysis experi-
ment (GEAE). The goal of the experiment is to compare a novel classification