Agriculture Reference
In-Depth Information
not in the Punjab (A.K. Singh, pers. comm.).
Also, although not frequently reported, repeated
plantings of wheat, barley, and oat cultivars with
a single gene for resistance to
H. avenae
have led
to selection of new virulent pathotypes over pro-
longed time periods, overcoming host-plant
resistance (Lasserre et al., 1996; Cook and Noel
2002), in addition to possibly increasing damage
from root-lesion nematode (Lasserre et al.,
1994).
It is also possible to manage damage by rotating
resistant cereals with susceptible crop species.
However, local knowledge of resistance reactions
is essential for effective use of this practice. For
instance, rye (
Secale cereale
L.) and certain culti-
vars of triticale (
Triticosecale rimpaui
Wittm.)
are resistant. Oat is resistant to
H. avenae
in
Australia and several Mediterranean countries
but susceptible in northern Europe (McDonald
and Nicol 2005). Moreover, resistant cultivars
from one region may be exposed to mixtures
of species in other regions, as exemplifi ed in
Israel by oat cultivars that are resistant to
H.
avenae
and susceptible to
H. latipons
(Mor et al.,
1992).
Host resistance will continue to be the most
profi table and easily applied management proce-
dure. However, resistance will only be used by
farmers if the cultivars also contain a level of tol-
erance (yield performance) which is comparable
to other commonly cultivated wheat cultivars.
Sources of resistance to
H. avenae
populations
worldwide have been collated and reviewed and,
where possible, have had their genetic location
and gene designation reported (Table 8.1)
(Rivoal et al., 2001; Nicol 2002; Nicol et al., 2003;
McDonald and Nicol 2005; Nicol and Rivoal
2007). All of the sources of resistance reported
against cereal cyst nematode to date feature single-
gene inheritance. Six
Cre
genes for
H. avenae
resistance in wheat (
Cre2
to
Cre7
) and the
Rkn2
gene for resistance to both
H. avenae
and
Meloido-
gyne naasi
(Jahier et al., 1998) were derived
from
Aegilops
species. Other resistance genes
were derived from
Triticum aestivum
(
Cre1
and
Cre8
) and
Secale cereale
(
CreR
). Several other
sources of resistance (
CreX
and
CreY
) are also
reported, but their genetic control and gene
designation are still unknown. Most of these
resistance genes have been introgressed into
hexaploid wheat.
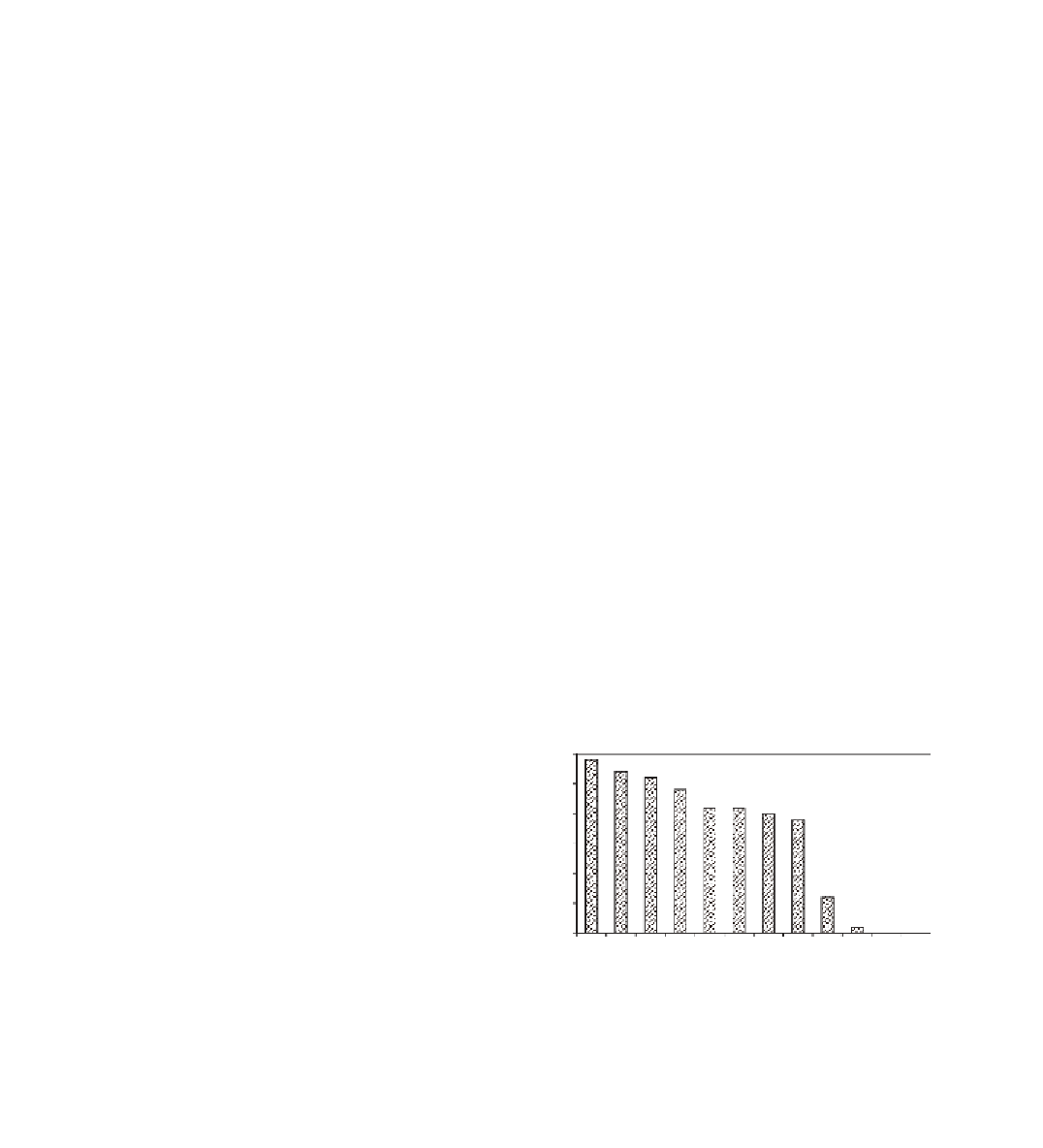
The
Cre1
gene is highly effective against popu-
lations of
H. avenae
from Europe, North Africa,
and North America (Fig. 8.1) and moderately
effective or ineffective against populations in
Australia and Asia (Rivoal et al., 2001; Mokabli et
al., 2002). Populations of
H. fi lipjevi
in India and
H. latipons
in Syria differ in virulence to the
Cre1
gene, compared with
H. avenae
(Mokabli et al.,
2002). In Turkey, the
Cre1
gene appears effective
against
H. fi lipjevi
, but
Cre3
is not. The
Cre3
gene
is effective against Australian populations
(Vanstone et al., 2008) but not European popula-
tions of
H. avenae
(de Majnik et al., 2003; Safari
et al., 2005) or
H. fi lipjevi
in Turkey. The
Cre2
and
Cre4
resistance genes from
Aegilops
and an
unidentifi ed resistance gene from the wheat line
AUS4930 offer promise against an array of
Heterodera
species and pathotypes (Nicol et al.,
2001). An International Root Disease Resistance
Nursery containing seven of the known
Cre
genes is coordinated by CIMMYT to establish
the value of these genes in different regions of the
world.
Molecular markers have been developed to
identify genes for resistance to
H. avenae
in barley
30
25
20
15
10
5
0
Fig. 8.1
Relative number of cysts for an Oregon population
of
Heterodera avenae
developing on root systems of 12
wheat cultivars or lines; the identity of a
Cre
resistance gene
is indicated if present.