Biology Reference
In-Depth Information
Ppy
+5
Ggo
(-5)
+5
Hsa
Ppa
+31
+10
(-3)
(-3)
+5
+5
+4
+9
Hla
(-5)
+2
Mfa
Mmu
+5
+1
+5
Atr
+5
+5
(-1)
+3
Cal
Cae
(-1, Ppy)
+1, Ppy
+10
(-1, Mfa)
+9, Mfa
+5
(-2)
+4
(-1)
+3
Cha
Soe
+12
+2
+3
+5
(-1, Cha)
+11
(-3, Atr, -3, Cal)
+6, Atr
+1
(-2, Guenons & Hsa)
+15
+7
+1, Cal
(-2, Guenons)
(-1, Cercopithecines)
5'
+10
3'
+1, Hla
Early region
Middle region & e/m extension
Late region
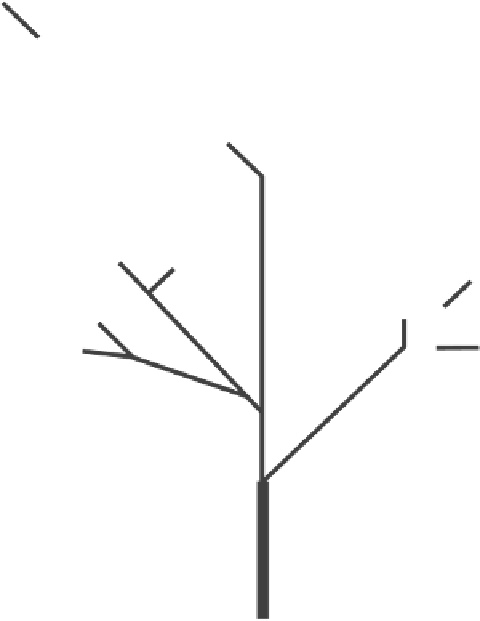
Figure 8.17.
Evolutionary tree of the M region of the primate involucrin gene detailing
repeat additions (after Green and Djian, 1992). The numbers along the branches of the
tree indicate the number of repeats added to form each region. Four late additions within
the middle or early regions and one 3
of the early region (boxed) are deviations from the
usual vectorial pattern of repeat addition. The deletions assigned to the region, the
lineage in which they occurred, and the number of repeats deleted are given in
parentheses. Ppy:
Pongo pygmaeus.
Hsa:
Homo sapiens.
Mmu:
Macaca mulatta.
Cal:
Cebus
albifrons.
Soe:
Saguinus oedipus.
Ppa:
Pan paniscus.
Hla:
Hylobates lar.
Cae:
Cercopithecus
aethiops.
Atr:
Aotus trivirgatus.
Ggo:
Gorilla gorilla.
Mfa:
Macaca fascicularis.
Cha:
Cercopithecus hamlyni.
glutamine-rich involucrin may have been able to function as a substrate for trans-
glutaminase.
Involucrin gene sequences have now been determined in a range of primate and
nonprimate species. Two different sites have been found within the coding region
that contain variable numbers of repeat segments. One, at site P, lies ~80 codons
from the ATG, contains a 16-amino acid repeat and is evident in nonprimate