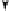Biology Reference
In-Depth Information
Potential acceptor site
Exon 3
Alu
Exon 4
OAT
gene
C
G
Donor site
OAT
mRNA:
splicing introduces
a new
Alu
exon
Exon 2
Exon 3
Exon 4
Exon 5
STOP
Figure 8.7.
Ornithine
-aminotransferase deficiency caused by a mutation in a resident
intronic
Alu
element within the
OAT
gene. A point mutation within the right subunit of
the inversely oriented intronic
Alu
repeat activates a donor splice site (from Labuda
et al
.,
1995).
Splice-mediated insertion of
Alu
sequences.
Alu
sequences have also been
found to alter protein coding sequences through the
splice-mediated insertion
of the
repeat and this probably represents the major mechanism by which
Alu
sequences
have entered protein coding regions.
Figure 8.7
illustrates the principle involved
by reference to the pathological example of ornithine
-aminotransferase defi-
ciency caused by a single base-pair substitution in an intronic
Alu
element in the
human ornithine
-aminotransferase (
OAT
; 10q26) gene; this lesion activates a
cryptic donor splice site which results in the incorporation of the
Alu
sequence
into the mRNA.
In an evolutionary context, there are several examples of the splice-mediated
insertion of
Alu
repeats into human gene coding sequences. The splice-mediated
insertion of a 95 bp
Alu
sequence has been reported in the lecithin: cholesterol
acyltransferase (
LCAT
; 16q22.1) gene (Miller and Zeller, 1997). In humans, the
alternate
Alu
-containing transcript represents between 5% and 20% of the
LCAT
mRNA. It is also present in
LCAT
mRNA from chimpanzee, gorilla and orang-
utan; in the latter, the
Alu
-containing mRNA species constitutes 50% of the total
LCAT
mRNA pool (Miller and Zeller, 1997). It is not however present in the
LCAT
genes of gibbons, or Old World and New World monkeys (Miller and
Zeller, 1997).
In the human biliary glycoprotein (
BGP
; 19q13.2) gene, three mRNA variants
are produced as a result of the alternative splicing of an exon (IIa) with one of two
virtually identical
Alu
cassettes derived from two intronic repeats (
Figure 8.8
).
Other such examples are to be found in the human
REL
(2p12-p13) proto-onco-
gene, and the complement decay-accelerating factor (
DAF
; 1q32) and comple-
ment C5 (
C5
; 9q33) genes (Makalowski
et al
., 1994). Of the 17
Alu
sequences
found in mRNA coding regions by Makalowski
et al
. (1994), seven contained in-
frame Stop codons and three others were predicted to cause frameshifts. Thus it is
perhaps not surprising that in several cases of mRNAs containing
Alu
sequences,
allelic exclusion
is evident and the mRNA containing the
Alu
sequence is of low
abundance compared to splice variants of the same gene that lack the repeat
(Mighell
et al
., 1997). This notwithstanding, it may well be that the splice-