Biology Reference
In-Depth Information
So far, the MHC appears to be confined to vertebrates (Klein
et al
., 1993; Klein
and Sato 1998). In a phylogenetic analysis of class I genes from different mam-
malian orders, Hughes and Nei (1989b) found that the class Ib genes clustered
with the class Ia genes. This is explicable either by postulating an independent
origin for the class Ib genes in different orders of mammals (Hughes, 1991) or by
invoking the homogenizing influence of gene conversion between class I loci
within each mammalian order (Rada
et al
., 1990). Class I genes evolved by a
process of repeated gene duplication followed by a reduction in the level of
expression of some genes, and the transcriptional silencing or deletion of others
(Watkins, 1995). This explains why orthologous relationships are found among
class I loci of mammals of the same order but not among mammals of different
orders. By contrast to the class I loci, orthologous relationships between mam-
malian class II loci are the rule rather than the exception, a consequence of the
early origin of the class II MHC loci prior to the mammalian radiation (Hughes
and Nei, 1990).
The MHC class I genes are conserved in the great apes and Old World monkeys.
Thus orthologues of
HLA-A
,
HLA-B
,
HLA-E
,
HLA-F,
and
HLA-G
are present in
macaques although the
HLA-C
locus has been found only in chimpanzees and
gorillas (Watkins 1995; Lienert and Parham, 1996). The MHC class I genes of New
World monkeys are similar to the
HLA-G
and
HLA-F
genes (Watkins, 1995). The
subfamily Callitrichinae (tamarins and marmosets) manifests an unusually high
rate of turnover of class I MHC loci with different sets of nonorthologous MHC
Present
b
a
B
C
E
c
L
J
A
d
K
H
G
e
F
b
a
B
C
E
c
L
J
A
H
G
d
K
e
F
b
a
B
C
E
c
L
J
AH
G
d
K
e
F
b
a
B
C
AH
E
c
L
J
G
d
K
e
F
ba
BC
AH
E
c
L
JG
de
KF
ba
BCAH
E
c
L
JGKF
de
ba
BCAH
E
c
LJGKF
de
ba
BCAH
ELJGKF
cde
MIC
HLA
BCAH
ELJGKF
bacde
300 Myrs ago
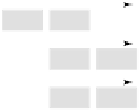
Figure 4.21.
Evolution of the class I MHC genes by serial duplication (after Klein
et al
.,
1998). Capital letters denote HLA loci, small letters MIC loci. Each rectangle represents
one locus. Multiple letters within a rectangle denote ancestors of genes specified by
individual letters. Short arrows indicate transcriptional orientation of loci whilst long
arrows denote transpositions of loci.