Biology Reference
In-Depth Information
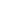
Gene duplication
Duplication of 22 or 11 codons
1
1
Deletion of 11 codons
6 or 7
1
1 or 0
1
2
1
3
APOA1
APOA4
APOE
APOA2
APOC3
APOC2
APOC1
Figure 4.6.
A hypothetical scheme for the evolution of the apolipoprotein genes (after
Luo
et al
., 1986). Numerals refer to the number of specified mutational events.
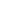
76
63
157
658
APOA1
134
127
1178
APOA4
44
66
193
860
APOE
34
76
133
230
APOA2
25
68
160
241
APOC2
36
68
124
308
APOC3
Figure 4.7.
Structural organization of the human apolipoprotein genes (after Li
et al
.,
1988). Exons are denoted by bars, the 5
and 3
flanking regions and introns by lines.
Open bars represent the 5
and 3
untranslated regions, hatched bars the signal peptide
regions and solid bars the mature peptide regions. The numbers above the exons indicate
their length in base-pairs.
The genes encoding the different components of the complement system
belong to several evolutionarily unrelated gene families. Phylogenetic analysis of
the
C3
,
C4
and
C5
genes, together with the more distantly related
2-macroglob-
ulin (
A2M
; 12p12-p13) gene, supports the view that C5 diverged first with C3 and
C4 subsequently diverging before the separation of the jawed and jawless fishes
(Hughes, 1994). Similar analyses have been performed for the complement C1
components (Dodds and Petry, 1993).
Exon shuffling has been extremely important in diversifying the structure of
the complement genes thereby conferring novel functions upon the complement
proteins. Thus for example, C1r, C1s, and C2 possess serine protease domains, C6,