Biomedical Engineering Reference
In-Depth Information
(JPSTH) for this type of analysis. The JPSTH has been used
to investigate relationships between neurons
(28)
and areas
(21)
. In many experiments, the same sequence of events
is typically repeated for several 'trials'. It is interesting to
know, then, how correlations between neurons relate to these
events. Essentially, the JPSTH analysis looks at time-varying
correlation between the neurons relative to stimuli or events
(
Fig. 7.5
).
To perform the JPST analysis, one should:
1. Arrange the spike trains for two neurons of interest into
perievent perievent matrices
; i.e., a matrix in which rows are
trials, and columns are bins (i.e., 25 ms) bins. Typically for
JPSTH analysis, one is not interested in fast interactions;
therefore, larger bin sizes (
10 ms) are preferred.
2. For each trial, record the bin in which spikes occurred for each
neuron. A Cartesian matrix can then be constructed in which
>
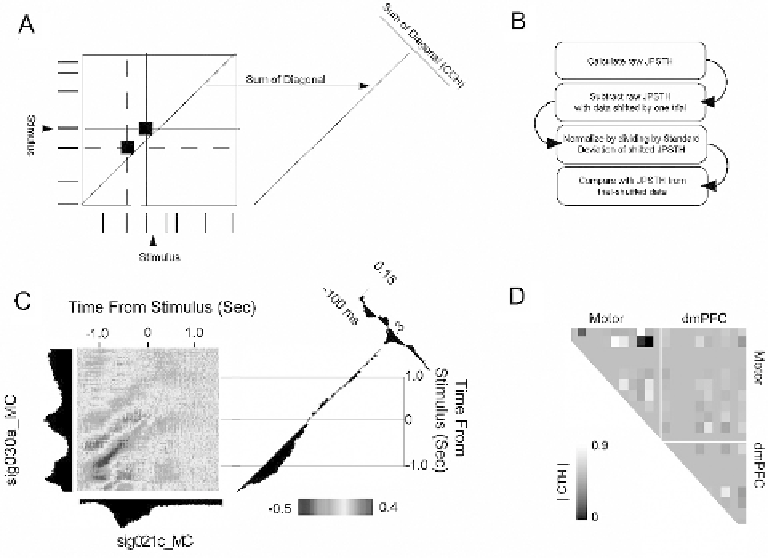
Fig. 7.5. Joint peristimulus time-histogram analysis. (
A
) For each trial, spike trains from one neuron are arranged on
one axis, and spikes from another neuron are arranged on the orthogonal (i.e., vertical) axis. Coincident spikes over the
course of the trial are recorded in the JPSTH matrix. This is repeated for every trial. (
B
) To correct for coincident spikes
related to simple modulations in firing rate, the shift predictor is subtracted from the raw JPSTH, and the entire quantity
is normalized to units of the shift predictor (by dividing by the standard deviation of the product of standard deviations of
PSTHs). Finally, the JPSTH matrix is compared with an identical matrix computed from trial-shuffled data. (
C
) JPSTH for
two motor cortex neurons reveals strong interactions prior to presentation of the stimulus. (
D
) JPSTH peaks across the
ensemble.