Image Processing Reference
In-Depth Information
Our image understanding system works in two modes: learning mode and classification
mode. In the learning phase, the system is supervised by an expert/physician. The task
here is to generate a set of fuzzy rules using fuzzy clustering and learning by examples
as described in Section 6.3.5 for which training data are required. In our case only input
values, e.g. object features such as object field, its shape coe
cient, from training data are
available. Outputs represented by class labels assigned to every object are generated during
clustering where the clustering results in the system learning phase should be additionally
accepted by the expert.
On the authority of histopathological findings (the “the gold standard”), contact endo-
scopic images were classified by pathologists into the following classes: tumor (SCC), severe
dysplasias (SD), mild dysplasias (MD). A control group, i.e. normal epithelium (NE) in-
cluded the images captured from unchanged epithelium. The results related to the four
specified histological classes are summarised in Table 6.6 and example images from the each
group are depicted in Fig. 6.9 as well as in (Tarnawski et al., 2008a).
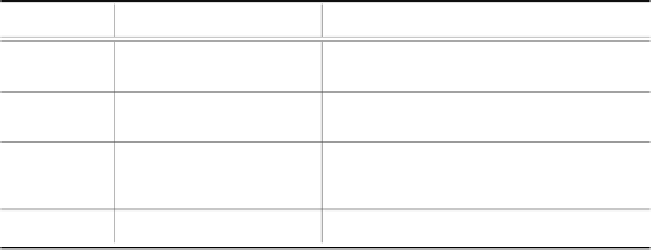
ImageType Histopathologicalresults Descriptionofthecontactendoscopyim-
agecharacteristics
SCC Squamous cell cancer
(FigX)
Disordered,crowdedcells,enlargednu-
clei,highvariablenuclearmorphology,
increasednucleustocytoplasmratio
SD Severedysplasia Enlargednuclei,crowdedcellswithmod-
eratevariablenuclearmorphology, in-
creasednucleustocytoplasmratio
MD Milddysplasia Moderatechangesinthesizeofthese-
lectednuclei,nucleispatialrelationships
slightlyabnormal,notuniformshapeof
thenuclei
NE Normalepithelium Regularcell formation,uniformround
nucleiwiththeslightsizevariation
TABLE 6.6
Description of CE images in the four analysed groups of histopathological findings.
For every group, the automatic method of cell nuclei detection described in (Tarnawski
and Kurzynski, 2007) was performed. Morphometric analysis was carried out on 26,260 cell
nuclei in total. Special software designed for the aim of this study helped to calculate 13
morphometric parameters for each nucleus:
•area and perimeter
•area to convex area ratio and perimeter to convex perimeter ratio
•length, width and length to width ratio (aspect ratio)
•elongateness coe
cient
•feret shape coe
cient
•blair-bliss shape coe
cient
•ratio of nucleus perimeter to the circle perimeter of the same size
•nuclei density index (based on the partition of the image space using multiple
grids of different size, also called multi-resolution grids). The value of this index
is defined as the combination of interpolated and weighted superposition of the
multi-resolution values of the nuclei density function. It is calculated for every
nucleus (for detailed description of this density function see (Tarnawski and Ci-
chosz, 2008)). This index takes decimal values from zero to one and describes
local nuclei distribution and density in the image.


Search WWH ::

Custom Search