Biomedical Engineering Reference
In-Depth Information
(a)
(b)
(c)
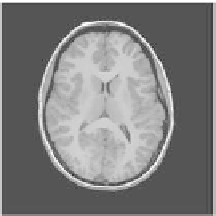
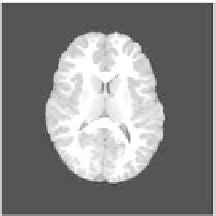
Figure 2.
Smoothing and skull stripping results using the BET2/FSL software: (a) slice
from original dataset: (b) smoothing results using the anisotropic filter: (c) skull stripping
result. The slice is from the savant dataset corresponding to a normal female, 25 years of age.
3.
IMAGE PROCESSING AND ANALYSIS
The data at hand undergo a series of preprocessing steps. First, all datasets
are smoothed using the anisotropic diffusion filter, which is known to preserve the
image edges. Then a brain extraction algorithm is applied to get rid of the non-
brain tissues in the savant images. Finally, all datasets are segmented into three
classes: white matter (WM), gray matter (GM), and cerebrospinal fluid (CSF).
3.1. Brain Extraction
Brain extraction, also known as skull stripping, is the process of removing
the non-brain tissues (e.g., skull, eyes, fat) from the MRI scans. The Brain
Extraction Tool (BET2) as implemented in the FSL package (free package at
http://www.fmrib.ox.ac.uk/fsl/) is used in this work to isolate the brain tissues and
get rid of all other artifacts. Figure 2 shows the result of applying the BET tool on
a T1-weighted MRI scan.
3.2. Image Segmentation
Three-dimensional segmentation of anatomical structures is very important
for various medical applications. Due to image noise and inhomogeneities, as
well as the complexity of anatomical structures, the segmentation process remains
a tedious and challenging process. Therefore, this process can not rely only on
image amounts of information, but has to involve prior knowledge of the shapes
and other properties of the objects of interest.
Deformable models have been extensively used for 3D image segmentation.
Such a model evolves iteratively toward the objects of interest by minimizing a
global energy. The functional energy represents the confluence of physics and