Biomedical Engineering Reference
In-Depth Information
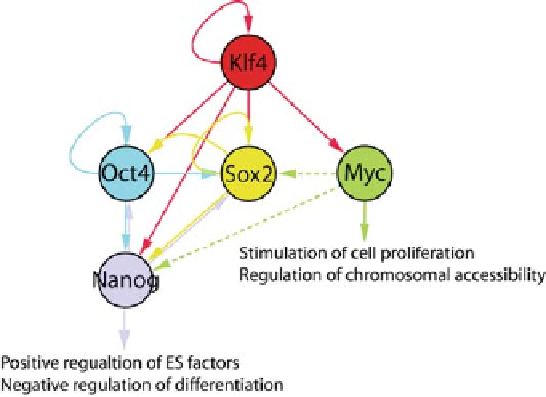
Fig. 3 Transcriptional regulatory circuit within four somatic cell reprogramming factors and
Nanog
factors (Chen et al., 2008). ChIP-seq is a variant of ChIP-ChIP that uses high-
throughput DNA sequencing rather than tiling arrays for detecting differences.
In spite of using different platforms, ChIP-ChIP and ChIP-seq, both reports
confirm an extended transcription factor network in mESCs. However, as
pointed out above, data have been accumulated by different platforms, such
as ChIP-ChIP, ChIP-PET, and ChIP-seq. Different platforms exhibit data
variations due to the technical differences in the methods, as well as in data
analysis. Comprehensive technological comparisons between different plat-
forms will be most useful in examining these networks in a comprehensive
manner (Euskirchen et al., 2007; Mathur et al., 2008).
miRNAs (microRNAs) are a class of short non-coding RNAs. They play a
crucial role of posttranscriptional gene regulation. miRNAs have been strongly
linked to stem cells, which have a remarkable dual role in development (Foshay
and Gallicano, 2007; Hatfield et al., 2005; Kanellopoulou et al., 2005; Murchison
et al., 2005; Shcherbata et al., 2006; Stadler and Ruohola-Baker, 2008; Wang
et al., 2007b; Yang et al., 2005; Zhang et al., 2006a). Marson et al. used ChIP-seq
methods to analyze how miRNA gene expression is controlled by key transcrip-
tional regulators in ESCs (Marson et al., 2008). Interestingly, genome-wide maps
of Oct4/Sox2/Nanog and Tcf3 (Cole et al., 2008) occupancy at miRNA promo-
ters are very similar to those of protein-coding genes. There are two sets of
miRNAs occupied by Oct4/Sox2/Nanog/Tcf3. One of them is actively expressed
in ESCs and the other is silenced in ESCs by polycomb genes in association with
the H3K27me3 mark. This result suggests that posttranscriptional control com-
plements transcriptional control in maintaining the pluripotent state.