Agriculture Reference
In-Depth Information
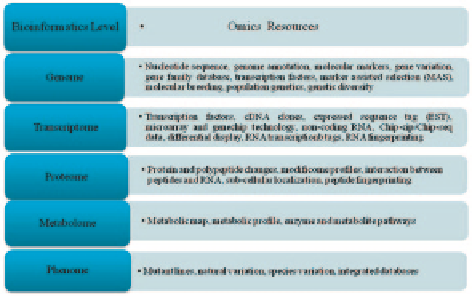
Fig. 2.1
A conceptual
model called 'omic space'
with layers ranging from the
'genome' to the 'phenome'.
(After Toyoda and Wada
2004
)
banks have been available for some time (Brady and Provart
2009
; Kuromori et al.
2009
; Seki and Shinozaki
2009
).
Bioinformatic information and web sites have become important for crop scien-
tists in gene data mining, and linking this knowledge to its biological significance
(Mochida and Shinozaki
2010
). However there needs to be a note of caution. As
genomic and proteomic knowledge expands, new forms of electronic data becomes
available to help interpret results. Biological data is notoriously variable (even unre-
liable at times) and 'noisy' in electronic form, due to living systems being complex
and measurement and analysis technologies are often imperfect. In my experience
two approaches for reducing 'noise' and help reliability of this type of data are
required; aggregation and visualisation. Firstly, when combined, multiple forms of
evidence become more and more accurate than for example a single source of data,
simply because each replicate form of the data reduces overall uncertainty. Sec-
ondly, the human mind is an outstanding data analysis tool. It can absorb textual
data rather poorly, but it can assimilate visual information in great detail, and the
mind can process visual data efficiently to help identify common trends and themes
(Cline and Kent
2009
).
In this chapter, we provide an overview of the many web-based resources avail-
able for use in 'omics' plant research, with particular emphasis on recent progress
related to crop species and crop improvement. Therefore we describe DNA and
RNA sequence-related resources, molecular markers, whole genome sequencing,
protein coding and non-coding transcripts, and provide molecular technology up-
dates. We then review resources important for genetic map-based approaches such
as QTL analyses and population genetic (diversity) studies. We also describe the
current status of resources and some technologies for transcriptomics, proteomics
and metabolomics; however some of these research areas are more comprehensive-
ly described in other chapters of this book. We then review molecular developments
in each 'omics' field, as well as instances of their combined uses in investigations
of particular crop systems. Mutant genotypes for use in 'phenome' research will be
discussed, and the integration of 'omics' data between plant species in comparative
genomics is dealt with. Throughout this review we provide examples of applica-
Search WWH ::

Custom Search