Agriculture Reference
In-Depth Information
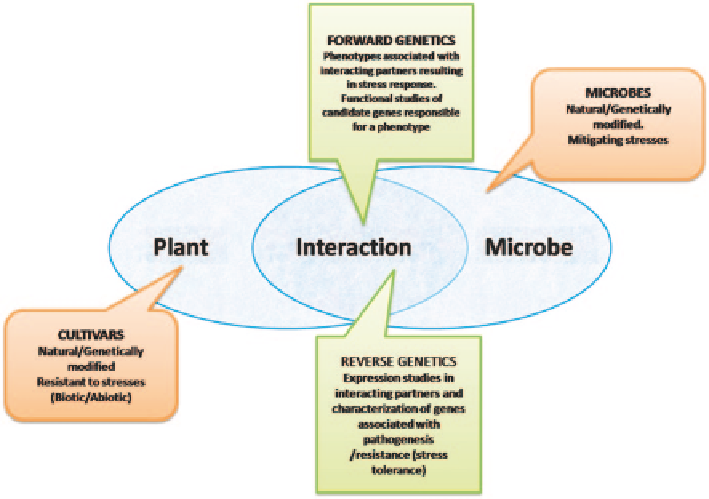
Fig. 8.1
Depicting approaches towards utilizing the interaction of plant-microbe for applications
in biotechnology. (Adapted from Schenk et al.
2012
)
revealing the mechanisms of antagonism between species infecting the same plant
as reported recently among Ustilago maydis and
Fusarium verticillioides
infecting
maize (Jonkers et al.
2012
). Based on the extensive collection of ESTs, several mi-
croarrays have been developed for crops (Torben et al.
2007
). The plant expression
database of plants and pathogens (PLEXdb, http://www.plexdb.org/index.php) can
provide the microarray data of
Fusarium graminearum
(obtained with Affymetrix
GeneChip) (Liu et al. 2010). The transcriptomics approach to study the interaction
between two- species may lead to the discovery of important genes in plants and
Fusarium
thus leading to characterization, which shall provide disease resistance
strategies (Fig.
8.1
). The multidisciplinary approaches are required for obtaining
a comprehensive systems biology look of the involved processes. The studies of
genomics, transcriptomics, proteomics and metabolomics will provide data sets,
which shall be integrated using bioinformatics and statistical tools and thus will
help to identify important biological processes and thus make prediction models.
These approaches are being used to access the information about the associations
(beneficial/detrimental) among two or multiple species. For example, the ectomy-
corrhizal interaction between
Laccaria bicolor
and aspen (
Populus tremuloides
)
roots led to the identification of genes expressed significantly, which were later
mapped to specific metabolic pathways and hence gave rise to the model of ectomy-
corrhizal metabolome. Identification of which genes are expressed is done by uti-
lizing the next-generation short-read transcriptomic sequencing data (Larsen et al.
Search WWH ::

Custom Search