Agriculture Reference
In-Depth Information
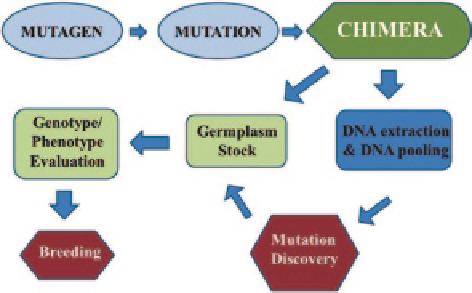
Fig. 4.6
A TILLING chart for gene function analysis and developing new crop varieties. A muta-
genized population is prepared using a mutagen that primarily causes small lesions (single nucleo-
tide polymorphisms, or insertions/deletions) randomly throughout the genome. Many mutagenic
treatments produce a chimeric plant in the first generation. Chimeras are dissolved and a structured
population is typically developed. A germplasm stock is prepared for long term storage of mutant
lines, and DNA is extracted from each individual mutant. DNAs are pooled and the library of
samples is screened for induced mutations in selected regions of target genes. Candidate mutants
are removed from the germplasm stock and further characterized genotypically and phenotypi-
cally. Individuals or lines exhibiting the desired characteristics can be incorporated into the breed-
ing program
because DHPLC will detect polymorphisms. Nevertheless, this strategy can be ap-
plied to species and hybrids that cannot be practically homozygosed: we and others
have detected rare polymorphisms in a heteroallelic background using DHPLC. The
general applicability of TILLING makes it appropriate for genetic modification of
crops, and there may be agricultural interest in producing phenotypic variants with-
out introducing foreign DNA of any type into a plant's genome.
TILLING consists of several major steps: development of a mutagenized popu-
lation, DNA preparation and pooling, and mutation discovery (Fig.
4.7
). At first,
random mutations are induced in genomes by using chemical mutagens; seeds are
mutagenized by treatment with ethyl methane sulfonate (EMS). The resulting M
1
plants are self-fertilized to get the M
2
individuals which are used to prepare DNA
samples for mutational screening. DNA is extracted from test samples. The DNA
samples are pooled and arrayed into microtiter plates. Screening for mutations be-
gins with PCR amplification of a target fragment of up to 1.5 kb using gene-specific
infra-red dye-labeled primers. The forward primer is 5′-end labeled with a fluores-
cent dye that is detected at 700 nm (IRDye 700) and the reverse primer is labeled
with the IRDye 800 nm (Till et al.
2006
). These PCR products are denatured and
re-annealed to allow the formation of mismatches, or heteroduplexes, which rep-
resent naturally occurring single nucleotide polymorphisms (SNPs) and induced
SNPs. Samples, are then incubated with a single-strand specific nuclease to digest
mismatched base pairs. For mismatch-specific cleavage, several enzymes, includ-
ing S1 nuclease (Howard et al.
1999
) and T4 endonuclease VII (Youil et al.
1996
)
Search WWH ::

Custom Search