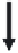Chemistry Reference
In-Depth Information
provides good support that the binding sites are co-localized and that binding is selective.
Frequently, it is observed that the small molecule is not displaced by the high-affinity ref-
erence compound. In these cases the project team may choose to de-prioritize such hits
or initiate other efforts to elucidate the biding mode. Our typical protocol is illustrated in
Figure 11.2.
L
1
L
2
L
3
Free ligand
A
NMR signals are negative
L
1
L
2
L
3
Add protein
L
1
NMR signals are
positive for bound
ligands
Bound ligand
B
L
3
L
2
Add high-
affinity
active-site
directed
competitor
NMR signal for
displaced ligand
becomes
negative again
C
L
1
Displaced
(free) ligand
L
3
L
2
Figure 11.2
Illustration of water-LOGSY NMR screening protocol. (A) Three representative
ligands (L
1
,L
2
, and L
3
) are combined in solution and show negative NMR signals for each
resonance in their spectra. (B) The target protein (large oval) is added to the mixture. Any
ligand that binds (in this case, L
1
and L
2
) shows a positive NMR signal. Those that do not bind
(L
3
) show no change in the NMR signal. (C) An active site-directed, high-affinity competitor is
added to the mixture. Any weaker ligand that occupied the active site is displaced back into
free solution and its NMR shift becomes negative again. Any ligand that is not displaced by
the competitor continues to have a positive shifted NMR signal. According to this example, it
would be suggested (but not proven) that L
1
is a competitive binder, the L
2
is a non-competitive
binder and L
3
does not bind.
To increase throughput, compounds were screened in groups of five in the presence of
BACE. If binding was detected for any of the compounds, each member in the set was
retested individually. The retesting process involved (1) analyzing the NMR spectrum of