Chemistry Reference
In-Depth Information
HIV integrase
10
4
Neuraminidase
10
3
HIV RT (nucleoside)
ICE1
PBP2x
PTP1b
IMPDH
Cathepsin K
ACE1
10
2
Factor X
HMG CoA reductase
10
DNA gyrase
Thrombin
Aldose reductase
EGFR
1
fCyp 51
HIV protease
Acetylcholinesterase
CDK
Enoyl reductase
PDE-4D
p38 kinase
0.1
Mdm2/p53
PDE-5
COX-2
0.01
cAbl kinase
HIV RT (NNRTI)
Druggable
Prodrug/transporter
Difficult
Undruggable
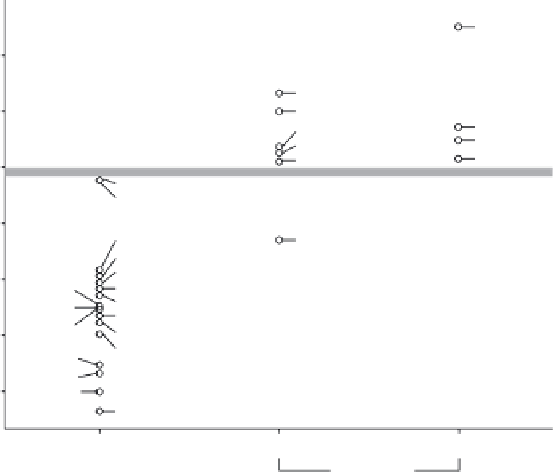
Figure 2.2
Calculated druggability for a set of 27 target binding sites. Known druggable
protein targets are shown on the left vertical, whereas known difficult targets (prodrug and
'undruggable') are shown in the right verticals. Difficult and druggable target binding sites are
effectively separated by the gray bar. The predicted druggability is the MAPpod score calculated
from the protein-ligand binding site structure. HMG-CoA, 3-hydroxy-3-methylglutaryl-CoA;
EGFR, epidermal growth factor receptor kinase; CDK, cyclin-dependent kinase 2; PDE,
phosphodiesterase; COX, cyclooxygenase; HIV RT, HIV reverse transcriptase; PBP2x, peni-
cillin binding protein 2x; IMPDH, inosine ICE1, interleukin-1 converting enzyme 1; PTP1b,
phosphotyrosine phosphatase 1b.
[28]
Reprinted by permission from Macmillan Publishers Ltd.
Inhibit
Screen small molecules
Discover Target
Forward Chemical
Genomics
change
Phenotype
Phenotype
change
Reverse Chemical
Genomics
Phenotype
Phenotype
Discover Target
Inhibit
Target
Figure 2.3
Diagram representing forward and reverse chemical genomic paradigms.
[32]
an observable change in phenotype so that a target may be discovered. Small molecules
interact with and alter the target and changes in phenotype are explored to connect it to the
pathway of interest. Forward chemical genomics can be described as screening a ligand
for a target. In reverse chemical genomics, targets without a known biochemical activity