Chemistry Reference
In-Depth Information
1
Pl
Pl
Pl
Pl
Pl
P
l
P
l
Pl
Pl
Y
Pl
Pl
749/822 (91%)
N
Y
2
N[
,
]
581/
701 (83%)
N
3
[N,O]
Y
CC
153/185 (83%)
N
Y
4
CX
198/300 (66%)
N
5
A
A
A
NH
2
A
Y
A
161/257 (63%)
N
6
H
1
Y
[N,O]
[N,O]
78/111 (70%)
N
Nonmutagen
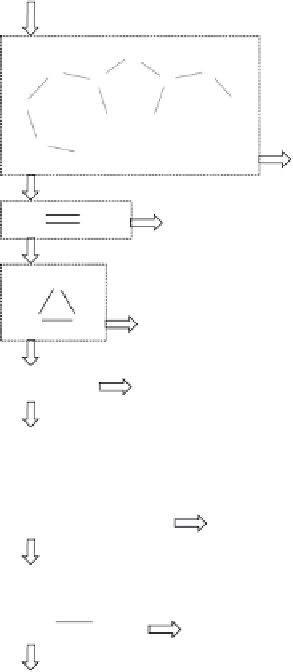
Figure 8.10
Decision list derived from mutagenicity data.
[40]
Arrows indicate the direction to
follow if a substructure is
(
Y
)
or is not
(
N
)
present in a compound. The number of mutagens,
the total number of compounds and the percentage of mutagens is indicated for each subset
(right).
pyridine, pyrimidine and pyrrole comprise almost 90% of monocyclic ring occurrences in
the fragmented data set. In addition, eight of the most abundant linkers were responsible
for 90% of all linker occurrences in the set. From the four rings and eight linkers, a virtual
library of kinase inhibitor scaffolds was constructed. Fragments known to form a critical
interaction with the binding site of a kinase, served as a starting anchor. New scaffolds
were generated by linking one of the rings to the anchor fragment, using one of the linkers.
This was repeated for all ring-linker combinations and for each attachment point on the
rings and anchor fragment. The newly designed scaffolds were docked against their targets,
using the placement of the anchor fragment as constraint. A fit-based score was calculated
and the highest scoring scaffolds were clustered according to the connection point at the
anchor fragment. Using this method, the authors were able to reproduce the predominant
structural motifs for known kinase inhibitors. In addition, they were able to suggest a
number of alternative variations for these ligand cores.