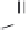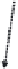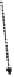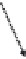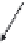Chemistry Reference
In-Depth Information
∗
N
C
O
C
C
O
C
C
O
N
C
O
O
C
C
C
C
C
C
C
C
C
O
C
N
O
O
O
O
C
C
C
C
C
C
C
O
C
C
O
C
C
C
N
C
C
C
N
O
N
O
C
O
C
O
C
C
O
C
C
C
C
C
C
N
O
C
O
N
N
O
C
C
C
O
N
Figure 8.4
The complete lattice of substructures for alanine (bottom). Level numbers in the
lattice increase with fragment size until the final structure of alanine is reached.
number of subgraphs at one level. Depth-first searching requires less storage, since a graph
is completely searched before advancing to the next. Therefore, it is proportional to the size
of the biggest graph. Modern graph mining algorithms, such as the ones described below,
work in a depth-first manner.
There are three problems central to frequent subgraph mining; the difference between
algorithms lies in how they address these problems. First, as was mentioned, subgraph
isomorphism tests are expensive in terms of computation steps needed to perform the
search. Second, the generation of refinements should be restricted. Third, since generated
duplicates require isomorphism tests, their number should be kept to a minimum, e.g. by
using a unique graph representation for testing.
The fragment miner MoFa (Molecule Fragment Miner)
[
7
]
was made especially for the
purpose of molecule mining. All embeddings are stored and used for isomorphism testing
and for restriction of fragment extensions to refinements that actually exist in the data-
base. To reduce the number of generated refinements, MoFa sorts all nodes and edges of
a fragment in the order in which they were added. Refinements may only occur at the