Chemistry Reference
In-Depth Information
5.8.2 CDK2 with Residue-specific Labeling
To assess how NOE matching will work using data from a residue-specific labeled protein,
the test case on the CDK2/
4
complex was re-run with modification of the NOE input list;
only those NOEs that could be observed in a residue-specific labeled sample were included.
While the development of cell free protein expression systems opens up all residue types
to selective labeling, we included only those residue types existing in the active site that
are routinely labeled in proteins expressed in
E. coli
. For the active site of CDK2, these
residues included isoleucine, valine, leucine, lysine, phenylalanine and alanine. NOEs from
residues such as aspartate and glutamate were removed from the list for this simulation. All
other parameters for the NOE run were as described above. The simulated NOE list for this
complex contained a total of 62 peaks, which were clustered into 40 protein
1
H
13
C groups.
The results obtained from applying NOE matching to CDK2/
4
are shown in Figure 5.18.
The posewith theminimumCOSTvalue has anRMSDof 0.92Å to the target pose. The pose
with the closest RMSD to the target pose itself ranks 19 out of 10 579 poses. In comparison,
if all NOEs are included as input for the NOE matching calculation, the pose with the
minimum COST value has an RMSD of 0.74 Å to the target (see the previous section).
(a)
(b)
6000
5000
4000
3000
2000
1000
0
0
1
2
3 4
RMSD to Target
5
6
7
8
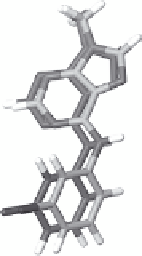
Figure 5.18
(A) COST versus the RMSD (Å) to the target pose for CDK2/
4
. The predicted
protein chemical shifts were set to the corresponding BMRB average values. The 3D X-filtered
NOESY spectrum was filtered to simulate data that could be extracted from residue-specific
labeled protein. (B) Superposition of target pose and the minimum cost pose (dark gray)
from (A).
5.9 Towards Larger Proteins by Nonuniform Labeling and
Stability Enhancement
In this section, some of the approaches described above for enhancing the sensitivity and
information content of protein-ligand NOEs are demonstrated for relatively large protein-
inhibitor complexes. In addition, we demonstrate that a medium-quality 3D X-filtered
NOESY spectrum can be obtained for a large protein-inhibitor complex by using a
stabilized, uniformly
13
C/
15
N-labeled protein sample in conjunctionwith an elevated experi-
mental temperature to increase the rotational correlation time of the protein-ligand complex.