Chemistry Reference
In-Depth Information
large and often poorly behaved proteins, we investigate the application of NOE matching
to these challenging systems. We describe how additional information can be obtained and
incorporated into the NOE matching protocol. With these enhancements, we expect that
NOE matching will enable the binding poses of ligands to be determined for a variety of
systems, including large complexes.
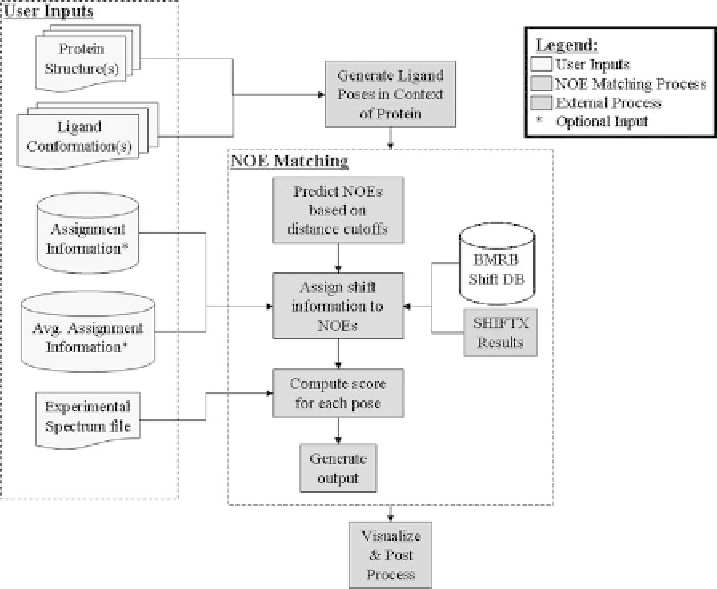
5.2 Summary of the NOE matching Protocol
The NOE matching protocol is described pictorially in Figure 5.1. Two files are needed
for input: an experimental NOE peak list with ligand protons assigned and a set of trial
binding poses to be evaluated or scored. The list of experimental peaks is typically derived
from a 3D
13
C-edited,
13
C/
15
N-filtered HSQC-NOESY spectrum.
[
16 20
]
(Hereafter, this type
of spectrum will be referred to as a 3D X-filtered NOESY spectrum.)
Figure 5.1
Flow scheme for NOE matching.
These peaks are grouped based on the protein
1
H and
13
C chemical shift values. This
procedure
identifies
(but does not
assign
)
1
H
13
C groups on the protein that give rise
to protein-ligand NOEs. Isotope-filtered NMR methods
[
19, 20
]
are applied to assign the
bound
1
H resonances of the ligand. (We note that for complex ligands, such as peptides and