Information Technology Reference
In-Depth Information
a.
0123456789012345601234567890123456
*+/+/a?a??a
4073
3704
09+++*+??a???737256
*+/+/a?a??a
09+++*+??a???737256
C = {1.227, 1.361, 0.512, 1.467, 1.666, 1.901, -1.583, 1.338, 0.620, -1.180}
C = {-0.286, -1.551, -1.165, 0.783, -1.872, 0.064, 1.152, -1.066, 1.343, 1.851}
1
2
b.
Sub-ET
1
Sub-ET
2
a
1.666
-1.066
0.783
a
a
a
1.227
1.338
-1.066
-1.165
0.064
c.
Sub-ET
1
Sub-ET
2
a
1.467
-1.066
0.783
a
a
a
-1.066
-1.165
1.338
1.227
0.064
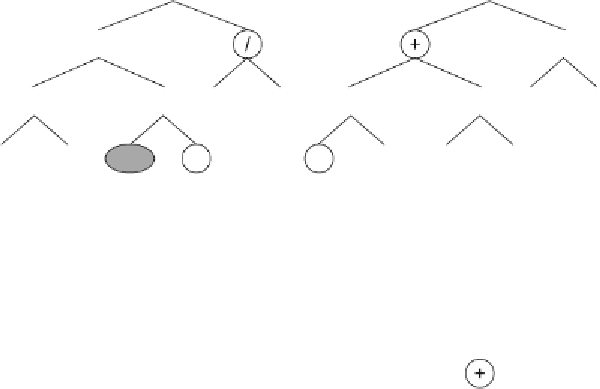
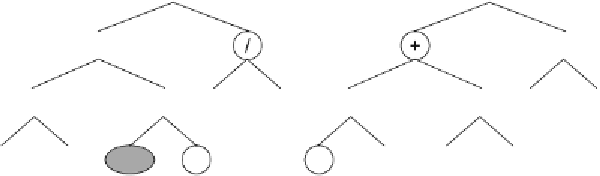
Figure 5.5.
Illustration of Dc-specific inversion.
a)
The mother and daughter
chromosomes with their random numerical constants (the RNCs are exactly the
same for both mother and daughter and therefore are shown just once).
b)
The sub-
ETs encoded by the mother chromosome (before inversion).
c)
The sub-ETs
encoded by the daughter chromosome (after inversion). The nodes affected by
inversion are highlighted. Note that not only a new constant is expressed in the
daughter tree but also different interactions are tested for two of the old constants.
Consider, for instance, the two-genic chromosome below with
h
= 5:
0123456789012345601234567890123456
+--+?aa?aaa655501-a+?-?aaa??577
250
(5.10)
and its arrays of random numerical constants: