Information Technology Reference
In-Depth Information
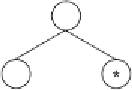
0.93
2.737
a
a
a
0.611
We will see in chapters 7 and 8 that genes encoding this type of domain
can be used to great advantage in polynomial induction and in parameter
optimization, as both these problems require large quantities or numerical
constants. But this elegant structure can also be used to design decision trees
with numeric attributes (see chapter 9) and to fine-tune the weights and thresh-
olds of evolvable neural networks (see chapter 10).
5.3 Multigenic Systems with RNCs
The creation of a multigenic system in which the basic units are the complex
genes of the previous section is very easy indeed, and can be achieved ex-
actly as we did for the basic gene expression algorithm (see section 2.1.3,
Multigenic Chromosomes). This means that each gene encodes a different
sub-ET and that the sub-ETs are posttranslationally linked by a linking func-
tion, creating a more complex program composed of multiple sub-ETs.
Consider, for example, the following chromosome with length 34, com-
posed of two genes, each with
h
= 5 and
n
max
= 2 and, therefore, with a gene
length of 17 (the Dc's are shown in bold):
0123456789012345601234567890123456
-//--?a?aa?
313500
/-*a-???a?a
185516
(5.5)
and its arrays of random numerical constants:
C
1
= {0.699, 0.887, -0.971, -1.785, 1.432, 0.287, -1.553, -1.135, 0.379, 0.229}
C
2
= {1.446, -0.842, -1.054, -1.168, 1.085, 1.470, -0.241, -0.496, 0.194, 0.302}
where C
1
represents the set of RNCs of gene 1 and C
2
represents the RNCs of
gene 2. This complex structure codes for two different sub-ETs, each