Information Technology Reference
In-Depth Information
In gene expression programming, UDFs may represent functions of sev-
eral variables although their arity in terms of expression rules is equal to zero.
That is, nodes with UDFs behave as leaf nodes or terminals. So, the imple-
mentation of UDFs in gene expression programming can be done using at
least two different methods: either they are treated as terminals and are used
both in the heads and tails of genes or they are treated as functions and are
used exclusively in the heads. Either way, the algorithm works very well.
Thus, it is a matter of taste which one to choose. I chose the latter as it seems
more consistent and less confusing. Consequently, UDFs are only used in
the heads of genes, they can occupy the first positions in the genes of the
individuals of the initial population, and they can also occupy the first posi-
tion of an RIS element.
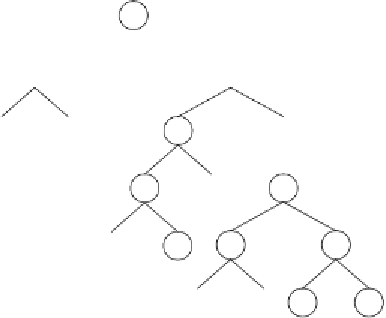
Let's now see how genes containing UDFs are expressed. Consider, for
instance, the chromosome below:
01234567890123456789012345678901234567890
AOOOdANUcAdOUcAAUOcddcbdaddaddcbdadbcdcbd
(4.25)
where “U” represents a user defined function. Its expression results in the
following expression tree:
A
O
O
O
d
A
N
c
U
A
d
O
c
A
A
U
c
U
O
d
c
d
This program encodes a perfect solution to the odd-4-parity function using
XOR as a UDF. The difference between this user defined XOR and a nor-
mal XOR is that the arguments to the user defined XOR are fixed during the
definition of the function, whereas the arguments to the normal XOR are
flexible and depend on the particular configuration on the expression tree. For