Information Technology Reference
In-Depth Information
otherwise stated, in all the experiments of this section, only mutation and two-
point recombination were used in order to simplify the analysis. The mutation
rate was equivalent to two one-point mutations per chromosome and the re-
combination rate was equal to 0.7. Furthermore, in all the experiments of this
section, the same set of 10 random fitness cases was used (see Table 4.2).
The fitness was evaluated by equation (3.3a) and, in this case, a selection
range of 100 and a precision of 0.01 were chosen. Thus, for a set of 10
fitness cases,
f
max
= 1000.
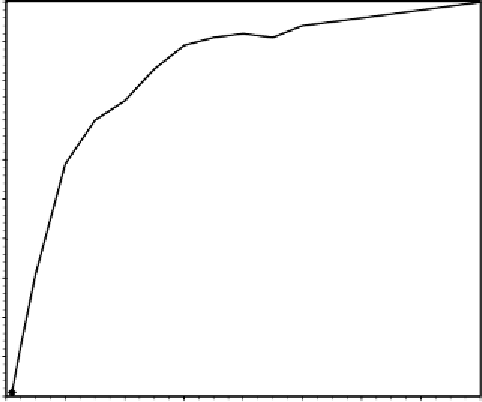
In the next experiment we are going to analyze the relationship between
success rate and population size
P
(Figure 4.2). This parameter is extremely
important not only in terms of evolution but also in terms of processing time.
And given the tradition (and necessity) of using huge population sizes in
genetic programming, it is important to show that the evolutionary system of
gene expression programming can evolve efficiently using small populations
sizes of just 20-100 individuals.
It is worth pointing out that, in this experiment, a medium, far from ideal,
value for the head (
h
= 19) was used in order to have some resolution in the
100
90
80
70
60
50
40
30
20
10
0
0
20
40
60
80
100
120
140
160
Population size
Figure 4.2.
Variation of success rate with population size in unigenic systems. For
this analysis,
G
= 50, and a medium, far from ideal, value of 39 for chromosome
length (
h
= 19) was used. The success rate was evaluated over 100 identical runs.