Biomedical Engineering Reference
In-Depth Information
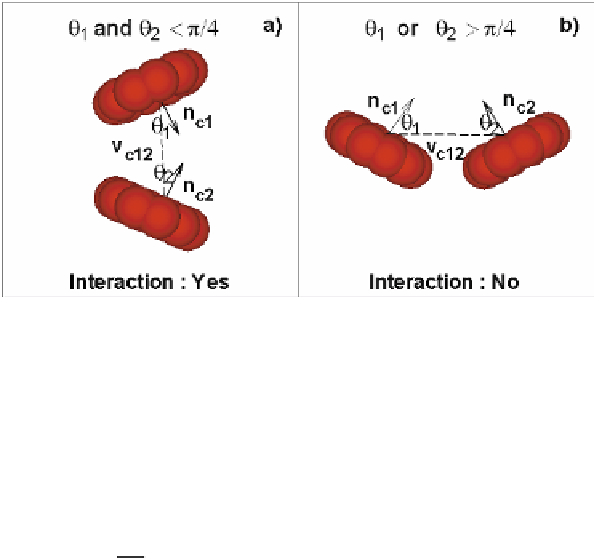
Fig. 10.6.
Schematic of the aggregation algorithm. Here, the two neighbor RBCs (1 and 2) are to
aggregate or not if the angles, θ
1
and θ
2
, are smaller or greater than π
/
4
where
r
M
is the model unit of length,
D
0
is the cell diameter, and
m
stands for meters.
The energy per unit mass (
k
B
T
) and the force unit (“
N
” denotes Newton) scales are
given by
Y
M
D
0
2
Y
P
Y
P
Y
M
D
0
D
0
M
P
N
M
N
P
(
k
B
T
)
=
(
k
B
T
)
,
=
,
(10.35)
D
0
where
Y
is the membrane Young's modulus. The time scale is defined as
D
0
D
0
P
Y
M
Y
P
s
η
τ
=
,
(10.36)
η
M
where
η
is a characteristic viscosity (e.g., solvent or membrane).
10.3 Parameter estimation
The models described in the previous section require as inputs “microscopic” pa-
rameters, e.g. the persistence length
p
for the WLC potential, but also other param-
eters, e.g. values of the membrane viscosity. These parameters may not be readily
available in the literature and certainly they vary according to the RBC state, i.e a
healthy or infected RBC. To this end, we aim to estimate most of the required param-
eters from single-cell measurements of macroscopic quantities, e.g. shear modulus,
which can then be mapped to “microscopic” (network) parameters using analytical
expressions, such as the one in Eq. (10.20). Specifically, the RBC model is compared
against several available experiments which examine cell mechanics, rheology, and
dynamics for healthy and diseased RBCs. First, we obtain the shear modulus us-
ing optical tweezers measurements of a stretched RBC. We then estimate the mem-
brane rheological parameters using measurements from optical magnetic twisting