Biomedical Engineering Reference
In-Depth Information
τ
(ms)
Coupled
G-S
Jacobi
0.25
0.50
1.00
1.25
1.50
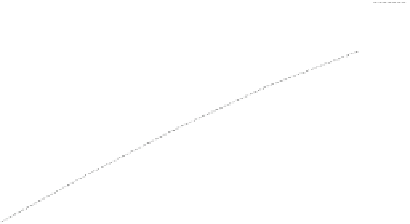
Fig. 4.7.
Top
: stability sensitivity to time-step size ( indicates numerical instability).
Bottom
: time
convergence history of the transmembrane potential error for Coupled (
u
e
,
V
m
)=(
u
n
+
1
e
V
n
+
1
m
(
,
)
),
u
e
,
V
m
)=(
u
e
,
V
n
+
1
m
u
e
,
V
m
)=(
u
e
,
V
m
)
Gauss-Seidel (
(
)
) and Jacobi
(
bidomain time-marching
schemes
Note that the cardiac subproblem (steps 1-3) can be solved independently of the
torso
subproblem
(step
4).
In
particular,
the
Gauss-Seidel-Robin
algorithm
u
e
,
V
m
)=(
V
n
+
1
m
u
e
,
V
m
)=(
u
e
,
u
e
,
V
m
)
(
)
, and the Jacobi-Robin algorithm
(
lead to
a fully decoupled computation of
w
n
+
1
,
V
n
+
1
m
,
u
n
+
1
e
and
u
n
+
1
T
. In other words, steps
1-4 are decoupled and can be performed sequentially.
u
e
,
V
m
)=(
u
e
,
V
n
+
1
m
u
e
,
V
m
)=(
u
e
,
V
m
)
Remark 8.
The choices
(
)
or
(
in Algorithm 2
allow for fully decoupled computation of
w
n
+
1
,
V
n
+
1
m
e
and
u
n
+
T
without the
need to resort to monodomain and uncoupling approximations (see Sect. 4.4).
,
u
n
+
1
Remark 9.
The interface coupling between steps 3 and 4 of Algorithm 2 corresponds
to the following Robin-Robin-based explicit time-discretization of (4.7):
t
T
t
T
+
γσ
+
γσ
u
n
+
1
u
n
+
1
u
T
·
u
T
σ
∇
n
=
−
σ
∇
n
T
·
on
Σ
,
e
e
e
T
h
h
(4.21)
t
T
t
T
n
T
+
γσ
n
T
+
γσ
u
n
+
T
·
u
n
+
1
T
u
T
·
u
n
+
1
e
σ
T
∇
=
σ
T
∇
on
Σ
.
h
h