Chemistry Reference
In-Depth Information
structure
property
name
TEXT
name
tvalue
nvalue
TEXT
cansmiles
coord
TEXT
NUMERIC[]
INTEGER
TEXT
NUMERIC
INTEGER
PK id
*
FK id
*
atom
INTEGER[]
sdf
molfile
TEXT
PF id
*
INTEGER
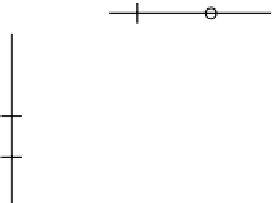
Figure 11.1
Entity relationship diagram for VLA4 schema.
example, SDF files were obtained from QSAR world,
3
a Web resource that
curates dozens of data sets used in quantitative structure activity relation-
ship (QSAR) studies. The VLA-4
4
Integrin antagonists were selected. This
file contains structures and data for 94 compounds.
5
One way to organize tables in a database is to define a new schema to
contain related tables. Here, we will create a schema name
vla4
. Using an
expansion of the example from the previous chapter, the following three
tables are suggested as a starting point. The entity relationship diagram
in Figure 11.1 illustrates the
vla4
schema.
Create Schema vla4;
Create Table vla4.sdf (id Integer, molfile Text);
Create Table vla4.structure (id Integer, name Text, cansmiles Text,
coord Float[][3], atom Integer[]);
Create Table vla4.property (id Integer, name Text, tvalue Text,
nvalue Numeric);
The column
structure.id
is a unique integer relating the structure,
sdf and property tables. The
sdf.molfile
column contains the mol-
file for each structure as defined by the vendor. The
structure.name
and
structure.cansmiles
columns contain the name and canonical
smiles parsed and computed from the molfile. The
structure.coord
column will contain an array of atomic coordinates. The
structure.
atom
column will contain an array of atom numbers from the file in
canonical order to correspond to the atom order in the canonical SMILES.
The OpenBabel/plpythonu extension functions
molfile _ mol
and
molfile _ properties
will be used to parse the vendor SDF molfiles
and populate these tables. The
molfile
column of the sdf table is first
populated from the SDF file, using the following perl script.




Search WWH ::

Custom Search