Information Technology Reference
In-Depth Information
Mixed Prediction and Prediction of Classifiers
Variational Bound and Number of Classifiers
200
8
data
pred +/- 1sd
gen. fn.
cl. 1
cl. 2
cl. 3
L(q)
K
1.5
7
150
6
1
5
100
4
0.5
3
0
50
2
1
-0.5
0
0
0
0.2
0.4
0.6
0.8
1
0
1000
2000
3000
4000
5000
Input x
MCMC step
(a)
(b)
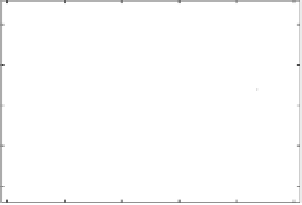
Fig. 8.6.
Plots showing the best discovered model structure for the generated function
using MCMC model structure search, and variational bound and number of classifiers
over the MCMC steps. Plot (a) shows the available data, the model of the classifiers,
and their mixed prediction with 1 standard deviation to either side, and additionally the
mean of the generating function. The matching function parameters of the classifiers
are
μ
1
=0
.
16
,σ
1
=0
.
01,
μ
2
=0
.
461
,σ
2
=0
.
025, and
μ
3
=0
.
78
,σ
3
=0
.
006. Plot (b)
shows the variational bound
L
(
q
) for each step of the MCMC algorithm, and clearly
visualises the random restarts after 500 steps. It also shows the number of classifiers
K
in the current model structure for each step of the MCMC search.
As can be seen in Fig. 8.6, MCMC model structure search performed better
than the GA by correctly identifying all 3 classifiers with
L
(
q
)
−
ln
K
!
≈
174
.
50,
indicating a higher
p
(
) than for the one found by the GA. While the dis-
covered model structure is not exactly that of the data-generating process, it is
intriguingly similar, given the rather crude search procedure. The reject rate of
the MCMC algorithm was about 96
.
9%, which shows that the algorithm quickly
finds a local optimum and remains there.
M|D
8.3.3
Sparse, Noisy Data
While the noise of the generated function is rather low and there is plenty of data
available, the next experiment investigates if the optimality criterion can handle
more noise and less data. For this purpose the test function from Waterhouse
et al. [227] is taken, where it was used to test the performance of the Bayesian
MoE model with a fixed model structure. The function is given by
f
(
x
)=
4
.
25(e
−x
4e
−
2
x
+3e
−
3
x
)+
4, and is shown in Fig. 8.7,
together with the 200 sampled observations. Waterhouse et al. used additive
noise with variance 0.44 which was here reduced to 0.2 as otherwise no pattern
was apparent in the data. It is assumed that the Bayesian MoE model was only
able to identify a good model despite the high noise due to its pre-determined
model structure.
Again using radial-basis function matching, the GA and MCMC settings
are the same as in the previous experiment, except for the initial number of
−
N
(0
,
0
.
2) over 0
≤
x
≤























































































































































































































































































































































































































































Search WWH ::

Custom Search