Information Technology Reference
In-Depth Information
M
K
m
nk
N
β
k
v
k
a
β
z
nk
x
n
b
β
α
k
W
k
a
α
y
n
b
α
data
a
τ
b
τ
τ
k
classifiers
Fig. 7.2.
Directed graphical model of the Bayesian LCS model. See the caption of
Fig. 4.1 for instructions on how to read this graph. Note that to train the model, both
the data
D
and the model structure
M
are assumed to be given. Hence, the
y
n
's and
M
are observed random variables, and the
x
n
's are constants.
specifies on one hand that
K
classifiers are
used, and on the other hand, where these classifiers are localised. Each classifier
k
has an associated matching function
m
k
:
The model structure
M
=
{
K,
M
}
[0
,
1], that returns for each input
the probability of classifier
k
matching this input, as described in Sect. 4.3.1.
Each input is assumed to be matched by at least one classifier, such that for
X→
each input
x
n
we have
k
m
k
(
x
n
)
>
0. This needs to be the case in order to be
able to model all of the inputs. As the model structure is known, all probability
distributions are implicitly conditional on
.
The data likelihood is specified from the generative point-of-view by assuming
that each observation was generated by one and only one classifier. Let
Z
=
M
z
n
}
be the
N
latent binary vectors
z
n
=(
z
n
1
,...,z
nK
)
T
of size
K
.Wehave
z
nk
=1if
classifier
k
generated observation
n
,and
z
nk
= 0 otherwise. As each observation
is generated by a single classifier, only a single element of each
z
n
is 1, and
all other elements are 0. Under the standard assumption of independent and
identically distributed data, that gives the likelihood
{
N
K
x
n
,
W
k
,τ
k
)
z
nk
,
p
(
Y
|
X
,
W
,
τ
,
Z
)=
p
(
y
n
|
(7.6)
n
=1
k
=1





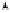












Search WWH ::

Custom Search