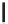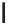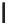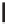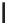Biology Reference
In-Depth Information
Allele T
G
C
A
A
G
T
A
C
C
T
A
Allele C
G
C
A
A
G
C
A
C
C
T
A
Figure 12.1
SNPs are created when the DNA replication enzymes make a mistake as they copy
the cell's DNA during meiosis. The enzyme incorporates the wrong nucleotide approximately once
every 2
−
3
×
10
8
bases. In the vast majority of cases SNPs are biallelic and only have two different
alleles. In the alleles shown above, the thymine nucleotide has been replaced by a cytosine. This is
a transition as a pyrimidine has been altered for another pyrimidine. A transversions occurs when
a pyrimidine (or purine) is change for a purine (pyrimidine)
of any given SNP, and this has been the major factor limiting their application to
forensic analysis: between 50 and 80 SNPs are required to achieve the same levels of
discrimination as the current STR-based methods [17, 18]. The technology used for
SNP detection is evolving and SNP analysis is becoming possible in many forensic
laboratories.
Detection of SNPs
There are many techniques available for the resolution of SNPs. In the 1970s it
was established that particular enzymes produced by bacteria can be used to cut the
DNA molecule by recognizing specific sequences [19]. Restriction digestion can be
used to genotype SNPs when the SNP either creates or destroys a particular restric-
tion enzyme recognition sequence [20, 21], but the method is limited for forensic
casework because it needs a large amount of DNA and is a long and laborious process.
Sanger sequencing
Sanger sequencing, also known as chain-termination sequencing, was developed in
the late 1970s and is a milestone in the development of molecular biology [22]. The
sequencing takes advantage of the biochemistry of DNA replication. The first stage
of the analysis is to amplify the target region using PCR; amplified products are
then used as the template in a sequencing reaction. The DNA sequencing reaction is
similar to PCR amplification and the reaction mixture is very similar, containing the
thermophilic
Ta q
DNA polymerase and deoxynucleotide triphosphates (dNTPs). It
differs from PCR in that only one primer is used and, in addition to the dNTPs, there
are four fluorescently labelled dideoxyribonucleotide triphosphates (ddNTPs); each
ddNTP is labelled with a different coloured dye [23]. The ddNTPs do not contain the
hydroxyl group on the 3
carbon, which prevents any extension of the DNA molecule
[24] (Figure 12.2).