Image Processing Reference
In-Depth Information
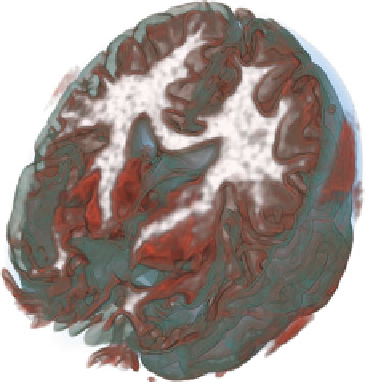
Fig. 1.4
A visualization of
the brain using transfer
functions that express the risk
associated with classification
More complex segmentation tasks cannot be achieved based on local image prop-
erties alone. They require models that account for more global assumptions or more
complex prior knowledge. Such models are also more computationally demanding
and are typically run as a pre-process of the visualization. Some of them output class
probabilities, from which Kniss et al. [
51
] derive measures that can be used to define
transfer functions that enable exploring the risk associated with binary classifica-
tions, or to visualize spatial decision boundaries. Figure
1.4
shows the use of such
transfer functions in a visualization of a segmented brain.
The framework of Saad et al. [
90
] combines volume rendering with tables that
list groups of voxels for which the same materials have been found to be most,
second most, and third most likely. They demonstrate several examples in which
these tuples can be used to detect anomalous subregions within areas that share the
most likely material. Follow-up work [
89
] has concentrated on identifying anomalies
or misclassification by considering regions in which the image-based likelihood
disagrees with shape and appearance priors.
Finally, work by Torsney-Weir et al. [
100
] addresses the model uncertainty in
segmentation methods by providing a systematic framework to explore the impact
of model parameters. This should facilitate finding settings that produce the desired
segmentation, and for which the results do not change significantly when slightly
changing the exact parameter values.
Fiber tracking, the reconstruction of nerve fiber bundles from diffusion MRI, is
another subfield of medical visualization in which uncertainty plays an important
role. It is treated in detail in Chap.
8
of this topic.

Search WWH ::

Custom Search