Image Processing Reference
In-Depth Information
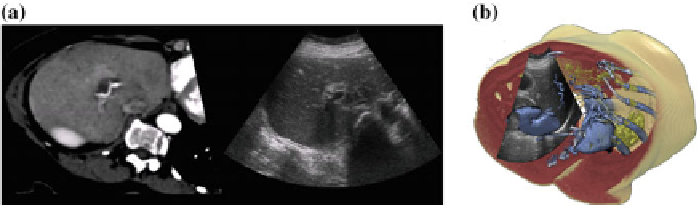
Fig. 24.5
Registering ultrasound and CT
a
: Slice-view of a CT scan co-registered with 2D ultra-
sound.
b
: Cut-away view of a CT scan co-registered with 2D ultrasound [
7
]
for instance sum-of-square-differences. Direct image based registration between ul-
trasound and CT or MRI can be difficult due to the different nature of the imaging
techniques and usually some pre-processing, such as filtering, is required. For in-
stance, an approach presented by Leroy et al. used a gradient-preserving speckle
filter and then looked for the similarity in the gradients.
Penney et al. proposed a technique for registeringMRI and ultrasound. The system
calculates a probability map of each element being a part of a liver-vessel [
52
]. Later
Penney et al. extended their technique for CT-ultrasound registration of the pelvis and
femur [
53
]. The system was validated using cadavers, showing that the registration
was accurate to a 1.6mm root-mean-square error on average. A similar technique
for the cardiovascular domain was proposed later by Zhang et al. [
83
].
Combining segmentation with registration, King et al. presented a technique for
registering pre-segmented models with ultrasound [
35
]. The technique predicts the
probability that the ultrasound image was produced by the segmented anatomy.
In addition to a rigid transformation, affine registration includes non-uniform scal-
ing which sometimes needs to be applied in order to get a more correct registration.
Wein et al. developed an automatic affine-registration technique between CT and
ultrasound [
78
]. To provide a better similarity of the ultrasound and CT, the system
creates a simulated ultrasound image out of the CT scan based on the tracked probe
position. The simulated ultrasound image is generated using a ray-traced approach
to calculate the ultrasound wave reflection and attenuation in the tissue. To simulate
tissue specific echogeneity, they apply an angle-independent polynomial function
based on which tissue the region corresponds to.
External pressure or different laying positions of the patient when acquiring the
images are influential factors. To account for local deformations while imaging soft
tissue, a more complex registration is required. Papenberg et al. proposed two ap-
proaches for CT ultrasound registration [
51
] given a set of paired landmarks in both
the CT and ultrasound data set. One approach uses the landmarks as hard constraints
and in the other, the landmarks are considered as soft constraints and are combined
with intensity value information, in this case the normalized gradient field. The pa-
per shows a non-rigid registration between the liver vascular structures. The latter
technique was later evaluated by Lange et al. [
40
].
Search WWH ::

Custom Search