Image Processing Reference
In-Depth Information
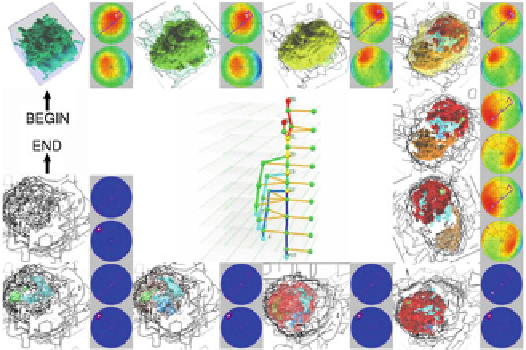
Fig. 23.2
Previewing a sheep heart volume along the optimal camera path. Image taken from
Ref. [
65
]
from the input medical datasets systematically and identify their spatial relationships
consistently. Actually, such topological concepts have been already introduced in the
earlier stage of medical visualization. For example, the 3D surface of a human cochlea
was reconstructed from a series of 2D CT cross-sectional images by identifying
correct correspondence between the cross-sectional contours [
62
].
Topological approaches have also been extended to analyze 3D medical volume
data. Contour trees [
2
] have been employed for designing transfer functions in order
to illuminate human organs and bones systematically since the associated anatomical
structure can be effectively captured as topological skeletons of isosurfaces [
75
]. Spa-
tial relationships between bones and the position of an aneurysm were successfully
extracted respectively from CT and angiographic datasets using a variant of con-
tour trees [
15
]. Interesting features in medical volume data can be visually analyzed
using an optimal camera path, which can be obtained by referring to the topolog-
ical structure of human organs [
65
] (see Fig.
23.2
). Topological methods are now
being developed for visualizing multi-variate and high-dimensional datasets, and
thus potentially for analyzing tensor fields obtained through DT-MRI, multi-subject
data in group fMRI studies, and time-varying data measured by high-speed CTs.
23.3.5 Integration of Simulation Models
In their 1999 predictive medicine paper, Taylor et al. argued that surgical planning
should not only address questions of surgical approach but also of the expected
outcome, for example, predicted future states such as the efficacy of a treatment
option or the performance of an implant [
66
]. Medical visualization approaches
become significantly more valuable when enhanced with simulation models that help
Search WWH ::

Custom Search