Biology Reference
In-Depth Information
Figure 3.2
Chart showing the approximate progression of peak supercomputing
capability (measured in floating point operations/second or instructions/
second) as a function of the year in which that capability became available.
of potential drug candidates against a particular receptor. These prob-
lems require large amounts of aggregate
capacity
, but for which the
individual calculations are of modest size. Grids and loosely coupled
networks of distributed workstations are examples of
capacity
systems.
Of course, any capability machine can also be used for capacity cal-
culations, but it is often not cost-effective to do so. Simulations of
large molecular systems that exhibit phenomena on long time scales
are
capability
calculations, but often one wishes to compute statistical
properties for which a number of separate trajectories may be used.
Also, some techniques require a number of
n
-body simulations to
be run in parallel with only loose coupling between them.
As the time scales accessible via simulation have become longer and
experimentally accessible time scales have become shorter, the ability
to connect simulation data to experiments in a significant way has
started to become a reality. This also allows the models (force fields)
used in simulation to be tested more extensively against experiment.
MOLECULAR SIMULATION
Molecular modeling and simulation by computer are techniques based
on the fundamental principles of physics. They have been used for many
years as a means to gain an understanding of materials and processes
at the molecular level and are capable of describing biological systems

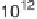
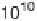

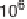
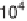





















































Search WWH ::

Custom Search