Biology Reference
In-Depth Information
Table 2.1 Preferred residue selection for positions 1, 4, 9, 10, 11, and 13 of
compstatin, as compared to the wild-type sequence
Position
Optimal
b
1 I A,V V,A
4 V Y,V W,Y,V
9 H T,F,A F,T
10 H H H,K,S
11 R T,V,A,F,H H,F,T
13 T V,A,F V,A,F
Only residues with greater than 10% representation among the lowest-lying energy
sequences are considered optimal. Provided in decreasing order.
a
Base case: positions 1 and 4 selected from {A,F,I,L,M,V,Y}; position 13 selected from
{A,F,I,L,M,V,Y,T}; positions 9,10, and 11 selected from all residues except C and W.
b
Base case with position 4 among {A,F,I,L,M,V,Y,W}.
Wild type
Optimal
a
Fold Specificity Calculations For Selected Sequences
Based on the sequence selection results, a handful of optimal sequences
were constructed for use in the second stage of the computational
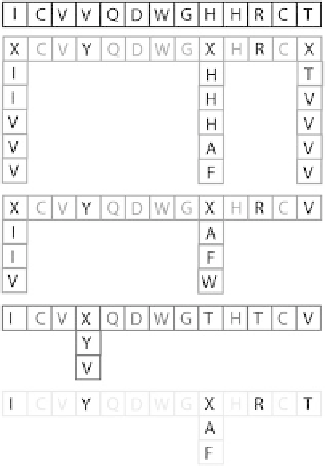
design procedure. Figure 2.1 presents the peptides studied which are
further classified into sets A, B, C, and D.
For all sequences further characterized via the fold stability calcula-
tions, residue 10 was set to histidine, a prediction consistent with the
Figure 2.1
Set of sequences tested for fold specificity.






Search WWH ::

Custom Search