Biology Reference
In-Depth Information
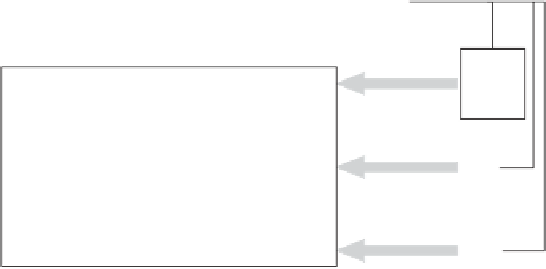
Figure 1.5
MudPIT platform is constituted of three major units: (i) sample
preparation, (ii) online liquid chromatography-tandem mass spectrometry,
and (iii) data analysis. (i) Protein samples are digested chemically with CNBr and
specific proteases (Endo-Lys C, trypsin) [27]. Alternatively, samples are digested
by a combination of nonspecific and specific proteases (elastase, subtilisin,
trypsin) and then pooled for LC-MS/MS analysis [28] or by a single nonspecific
protease (proteinase K) [80]. Using nonspecific proteases increases the
sequence coverage of posttranslationally modified regions of proteins [28, 80].
(ii) The sample is then loaded onto a narrow capillary column and peptides are
alternatively eluted from SCX and RP phases [26,27]. Peptides are ionized by ESI
and introduced in a tandem mass spectrometer. (iii) Data analysis is performed
by a suite of software packages. Peptides are identified by Sequest [18],
PepProbe [67], or GutenTag [122]. Peptide information is assembled in protein
identification [68]. Using DTASelect [78], results are further inspected based on
criteria of interest. Quantitative experiments using stable isotopically labeled
samples use Relex software for obtaining the relative protein abundances [59,60].
Statistical modeling of the data [57] or experimental parameters [77] can be used
to empirically estimate amounts of proteins without stable isotope labeling.
17



























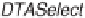













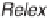





































Search WWH ::

Custom Search