Biology Reference
In-Depth Information
Current databases can only paint a rough sketch of a few well-studied
transcriptional pathways and more data needs to be generated to add
to the power of predictive models. Indeed, a recent genetic screen on the
Wnt
pathway in
Drosophila
suggests that some transcriptional pathways
may in fact involve many more molecular branches and connections
than expected [30].
IDENTIFICATION OF KEY TRANSCRIPTION FACTOR BINDING SITES IN PROMOTER
REGIONS OF ES GENES BY SEQUENCE COMPARISON
Several genes that appear to be key functional regulators of pluripo-
tency maintenance and differentiation have been identified in ES cells.
These regulators include the transcription factors
Oct4
,
Sox2
, and
Nanog
.
Oct4
binds to a specific DNA octamer, ATGCAAAT, within pro-
moter regions of target genes.
Oct4
is highly expressed in ES cells and
its reduction is necessary for differentiation to proceed [31-33].
Oct4
targets and controls the expression pattern of a number of genes (
Fgf4
,
Utf1
,
Opn
,
Rex1/Zfp42
,
Fbx15
), whose regulatory elements are found to
contain putative in silico
Oct4
binding sites. Mutation analysis of these
putative sites has confirmed their importance in modulating gene
expression via
Oct4
binding ex vivo. Interestingly, when computational
genome-wide scanning was performed in several species, a conserved
Oct4
octamer binding sequence was found to be present within the pro-
moter regions of
Oct4
,
Sox2
, and
Nanog
genes. The binding of
Oct4
to
these sites was confirmed by chromatin immunoprecipitation (ChIP)
[34]. These exciting findings (figure 10.5) demonstrated that these three
key ES cell transcription factors autoregulate each other via an intricate
network, where the molecular details have yet to be fully elucidated.
Figure 10.5
The network of genes regulated by two key ES transcriptional
factors. The feedback regulation of the transcriptional factors
Oct4/Pou5f1
and
Sox-2
are shown, interconnected by each binding to the other's promoter.
Oct4
and
Sox2
colocalized right next to each other in their own promoters and in
many target genes, examples of which are shown in the figure.

















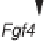














Search WWH ::

Custom Search