Biology Reference
In-Depth Information
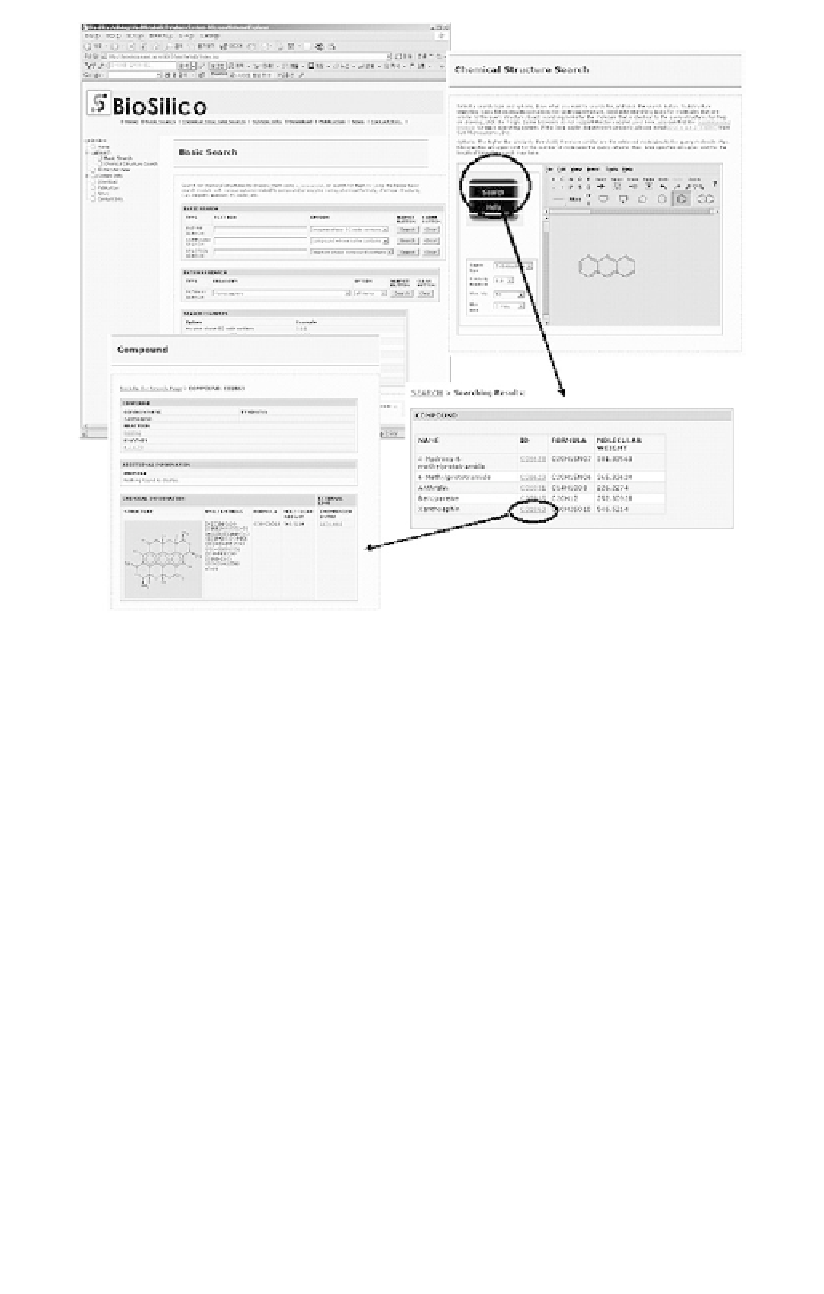
Figure 7.6
Database query interface of BioSilico.
This is indeed needed for researchers who are less familiar with the
computational methods in detail. For this purpose, a program package,
MetaFluxNet, was developed, which provides one of the systems
biology platforms for metabolic characterization and engineering [76].
M
ETA
F
LUX
N
ET
MetaFluxNet is a program package for managing information on the
metabolic reaction network and for quantitatively analyzing metabolic
fluxes in an interactive and customized way. Users can interpret
and examine metabolic behaviors and changes in response to genetic
and/or environmental modifications. Consequently, quantitative in silico
simulations of metabolic pathways can be carried out to understand
the metabolic status and to design the metabolic engineering strategies.
The main features of MetaFluxNet include a well-designed model
construction environment, a user-friendly interface for constraints-
based flux analysis, comparative flux analysis of different strains under
varying environmental conditions, several options for choosing numer-
ical solvers, and automated pathway layout creation [76].
Search WWH ::

Custom Search