Biology Reference
In-Depth Information
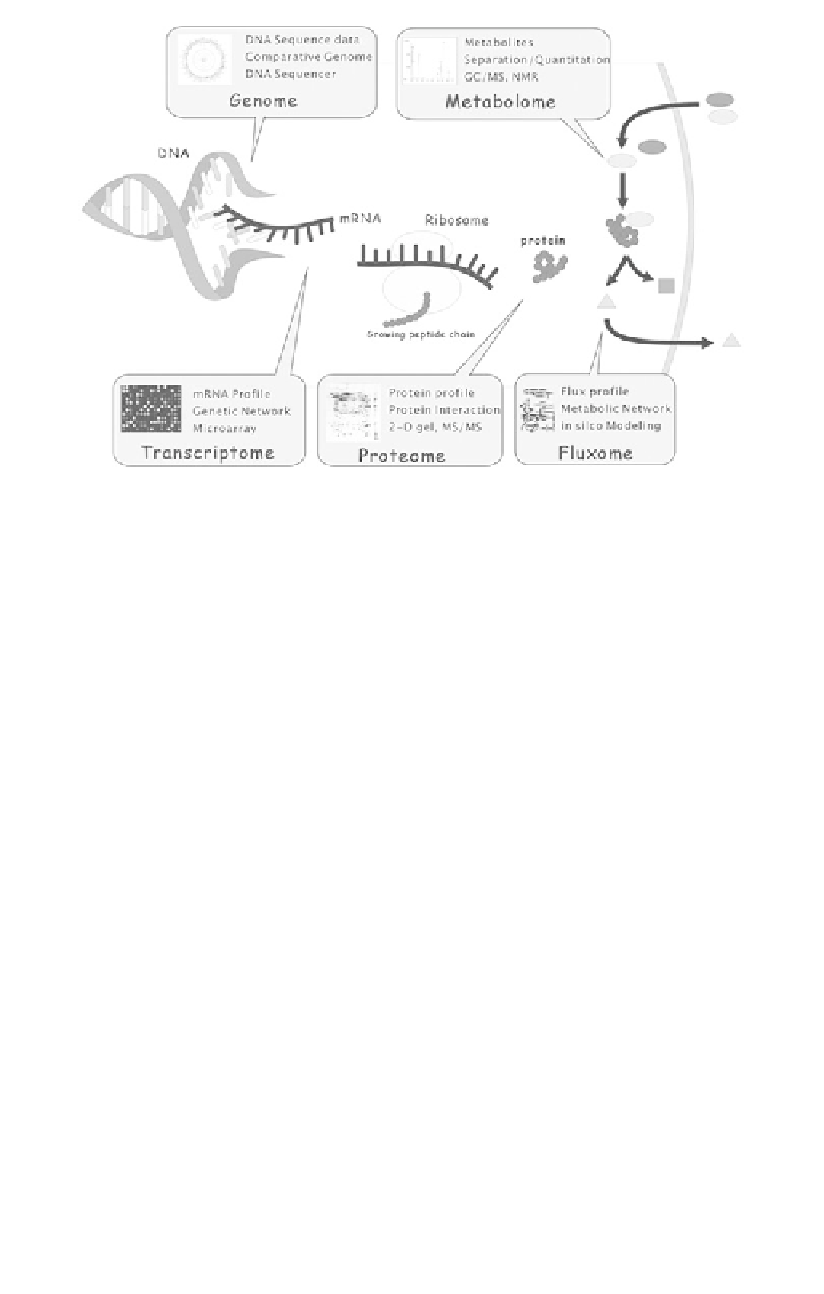
Figure 7.2
Postgenomic omics research. High-speed DNA sequencing has
allowed completion of a number of genome sequencing projects. Having complete
genome sequences for many microorganisms in our hands, comparative
genomic studies can be performed. Development of high-density DNA
microarray allowed profiling of the transcriptome of cells under investigation,
thus allowing us to examine the expression levels of multiple genes
simultaneously. Two-dimensional gel electrophoresis (2DE) combined with
mass spectrometry made it possible to obtain complete profiling of cellular or
subcellular proteins, which complements the results of transcriptome profiling.
Metabolome profiling is also becoming increasingly popular as better GC/MS,
LC/MS, and NMR techniques are developed. Fluxome profiling is still mostly
based on computation, but is supplemented with the results of isotopomer
profiling for better estimation. Much effort is being devoted to integrate all
these omics data toward better understanding of global cellular physiology,
metabolism, and regulation, and consequently to use the results for strain
improvement.
activities of these enzymes. Metabolome profiling can complement
some of these limitations, but is currently hampered by the absence
of robust methods for sample preparation and metabolites analysis.
Complementary to this, fluxome profiling allows determination of
metabolic fluxes by metabolic reaction network simulation constrained
by the measurement of net excretion rates of extracellular metabolites,
uptake rates of metabolites, and/or isotopomer distributions following
the use of isotopically labeled substrates.
Search WWH ::

Custom Search