Biology Reference
In-Depth Information
Concurrently with high-throughput experiments, it is increasingly
accepted that in silico or dry experiments of modeling and simulation
improve our ability to analyze and predict the cellular behavior of
microorganisms under any perturbations, for example, the genetic
modifications and/or environmental changes [18-20]. Thus, metabolic
engineering of microorganisms can be best performed by integrating
wet experiments (high-throughput experiments) and dry experiments
(in silico modeling and simulation) at the systems level (figure 7.1);
the results of genomic, transcriptomic, proteomic, metabolomic, and
fluxomic studies, information and data available in public and in-house
databases, and those generated by in silico analysis can be integrated
within the global context of the metabolic system. This leads to the
generation of new knowledge that can facilitate strain development
suitable for biotechnological processes with the highest efficiencies
and productivities. Hereafter, such a systems approach is referred to
Figure 7.1
Overview of systems biotechnology. High-throughput experiments
generating large amounts of genome, transcriptome, proteome, metabolome,
and fluxome data can be analyzed in combination with in silico modeling and
simulation results in order to develop improved organisms for applications in
biotechnology industries.








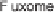














































Search WWH ::

Custom Search