Biology Reference
In-Depth Information
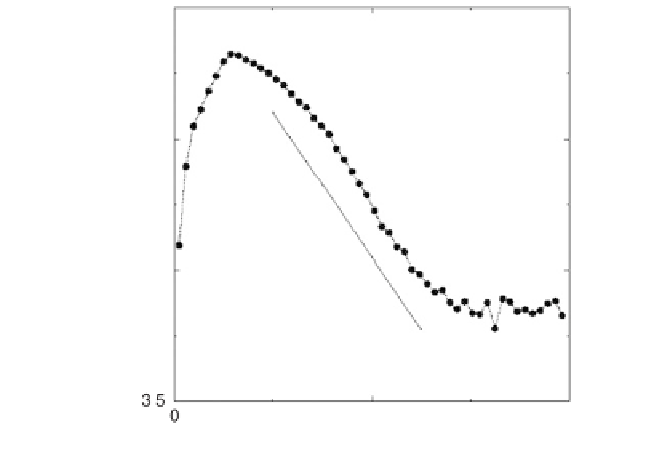
Figure 4.5
Logarithm of the hybridization noise versus the expression level for
DNA microarray data.
It is noteworthy that calculation of the hybridization noise strength
s
hyb
for nine pairs of replicate experiments [15], which were performed
with a different type of Affymetrix GeneChip array (HuGeneFL),
with a different type of cell (human fibroblast cells), and in a different
laboratory, yielded an exponentially decaying component of the
hybridization noise the same as that observed above [12]. That is,
the noise as characterized in the present analysis seems to show a
degree of universality, although more work will be needed to confirm
this behavior.
USE
-
FOLD
:
A METHOD FOR UNIFORM SIGNIFICANCE OF EXPRESSON
FOLD CHANGE
The results presented in the previous subsections can be used to design
a method for determining the statistical relevance of gene expression
changes. The idea is simply that the fold change experienced by a gene
under different biological conditions has to be larger than the fold
change expected from the noise. The noise distribution function
discussed above can be used to evaluate the significance of the differ-
ence between a pair of gene expressions (q
1
,
q
2
) for the same gene
but from different experiments. One begins with the null hypothesis
that the q
1
and q
2
arise from the same distribution (
G
2
) and that the
difference between them is due to noise. One then defines a gene
expression dependent
p-
value as

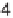
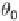


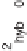

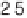











Search WWH ::

Custom Search