Biology Reference
In-Depth Information
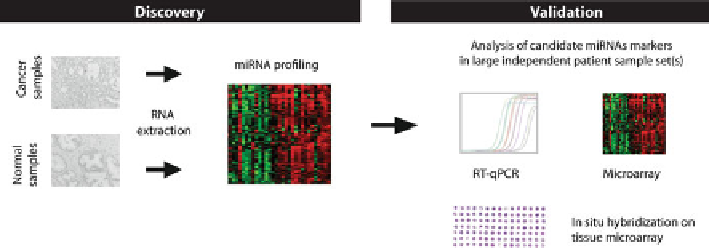
Fig. 13.2
Strategy for identification of miRNA biomarkers in cancer
. MicroRNA expression
profiling of patient samples (normal vs
.
cancer tissue samples shown here) is performed to identify
candidate miRNA markers for a given malignancy. Microarray analysis is commonly used in the
discovery phase, but other profiling methods are also available (see Fig.
13.3
). Novel candidate
miRNA markers identified in the discovery phase are subsequently validated in large independent
patient sample set(s) by RT-qPCR or in some cases by microarray analysis or in situ hybridization
using tissue microarrays. Only a subset of miRNAs will pass validation
13.2.2
Technologies for miRNA Pro fi ling
Several technologies are available for determination of miRNA expression levels
and can be divided into three overall types: hybridization-based [DNA microarrays,
in situ hybridization (ISH), Northern blotting], amplification-based [reverse tran-
scription quantitative PCR (RT-qPCR)], and sequencing-based [next-generation
sequencing (NGS)] [
90,
91
]. Using DNA microarrays, it is possible to measure the
expression of hundreds of miRNAs in parallel in a single experiment [
92,
93
] .
Typically, fluorescently labeled RNA (or reverse-transcribed cDNA) molecules are
hybridized through Watson-Crick base pairing to complementary DNA oligonucle-
otides (probes) immobilized on a microarray glass (or quartz) slide (Fig.
13.3
, left
panel). The position of each type of probe on the microarray is predefined (referred
to as a “spot”), and the intensity of the fluorescent signal from a given spot after
hybridization is taken as a measure for the expression level of the corresponding
miRNA in the sample analyzed [
94,
95
] . Today, several commercial DNA microarray
platforms are available for relatively cost-effective and user-friendly profiling of
miRNAs [
96-
99
] .
Although intra-platform concordance is generally high, inter-platform variance
can be substantial because of differences in exact microarray designs (e.g., probe
type, length, and sequence) and experimental protocols [
99-
102
] . For example,
some protocols use total RNA as input (includes mature, pri-, and pre-miRNAs),
while others employ an enrichment step for small RNAs (mainly mature miRNAs)
prior to labeling and hybridization. These approaches will produce different results,
since probes directed at the mature miRNAs sequence will also recognize the cor-
responding pri- and pre-miRNA. Furthermore, the short length (~22-nt) of mature
miRNAs, many of which vary in sequence by only 1 or 2 nt, constrains probe design,
and cross-hybridization to closely related miRNAs remains a significant challenge
Search WWH ::

Custom Search