Biology Reference
In-Depth Information
nearly all changes in RAPD profiles were reversible after 8 or 12 days of recovery, which
suggests that
Daphnia
can repair most of the DNA lesions. The changes in RAPD profiles
were more pronounced in moribund individuals compared to active swimming
Daphnia
exposed to B(
a
)P. Some of the changes were transmitted from the B(
a
)P-exposed adult
Daphnia
to the neonates (first two generations). It is noteworthy that none of the neonates
was exposed directly to B(
a
)P. This suggests that genomic alterations occurred in germi-
nal cells in adult
Daphnia
and were transmitted to the next generations (Atienzar and Jha
2004). These results illustrate well the power of the RAPD approach to detect genomic
changes after careful optimization.
Similar approaches have been used to study the effect of genotoxins on gene expres-
sion. The techniques developed without any knowledge of the genome are: RNA-
arbitrarily primed polymerase chain reaction (RAP-PCR; Welsh et al. 1992), subtractive
hybridization techniques (Sargent and Dawid 1983), and differential display (Liang and
Pardee 1992). For instance, Rodius et al. (2002, 2004) performed RAP-PCR to study the
impact of aquatic pollutants on the transcriptome of freshwater bivalves. This approach
uses random primers to amplify cDNA, then the amplimers are separated by electro-
phoresis on agarose gel, and the expression patterns of organisms from polluted sites
are compared to controls. Freshwater mussels (
U. tumidus
) from a control area were
transferred for 21 days to two sites in the Moselle River, upstream and downstream of
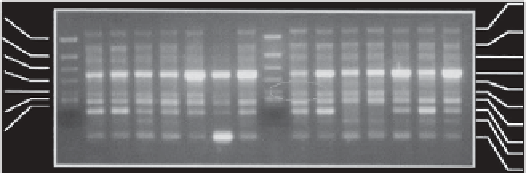
urban and industrial areas (Rodius et al. 2004). An additional amplimer was observed
in the pattern of the bivalves transplanted to the downstream site compared to controls
(Figure 13.3).
This amplimer has been cloned and sequenced. PCR performed with specific primers
showed that this product is not the result of a polymorphism but corresponds to gene
induction in the transplanted mussels. However, the analysis of about 40 mineral and
organic pollutants in the sediments did not allow the identification of pollutants which
could explain the modification of the expression pattern. Moreover, the amplimer coding
and translated sequences did not show any homology with nucleotide or protein data-
bases at the National Center for Biotechnology Information site, so no function concerning
this gene could be suggested.
Considering that the sequencing of the genome of aquatic species is becoming increas-
ingly accessible, the field of ecotoxicogenomics is expected to progress in the coming years
(Lettieri 2006; Piña and Barata 2011). This will help us understand the observed responses
to environmental stressors in contaminated environments (Calzolai et al. 2007).
bp
1057
770
612
495
392
340
294
M C1C2 C3 C4 C5 C6 C7 M
F1 F2 F3
F4
F5 F6 F7
1
2
3
4
5
6
7
10
PCR
products
FIGURE 13.3
Comparison of RAP-PCR amplification patterns of mRNA from digestive glands of
Unio tumidus
using the AP1
primer. PCR products (amplimers) are numbered to facilitate pattern description. Mussels have been trans-
planted at C (control) and F (polluted) sites. M, DNA size marker.

Search WWH ::

Custom Search