Biology Reference
In-Depth Information
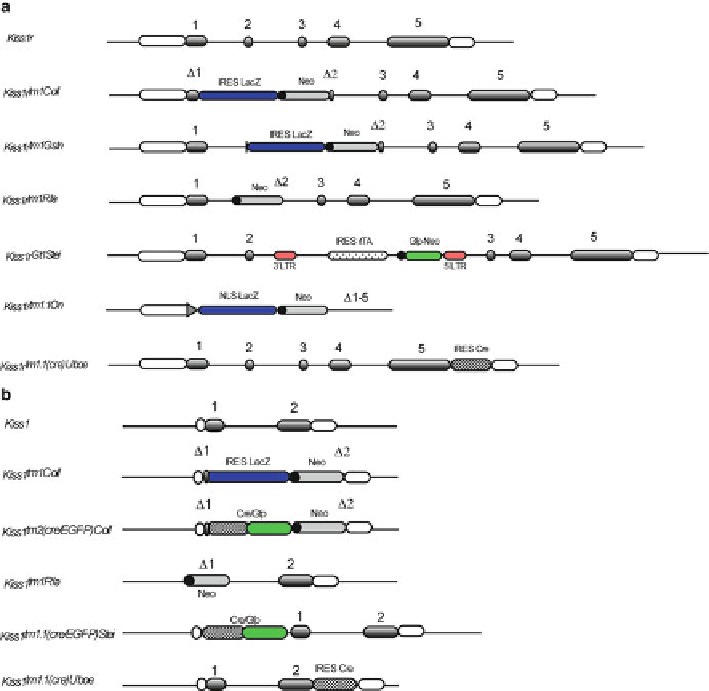
Fig. 22.1
Genomic arrangement of
Kiss1r
and
Kiss1
targetted loci. The nomenclature for these
transgenic lines is based on the guidelines established by the International Committee on
Standardized Genetic Nomenclature for mice (
http://www.informatics.jax.org/mgihome/nomen/
index.shtml
)
and follows that used on the Mouse Genome Informatics website (
http://www.infor-
matics.jax.org/
). Lab Codes are: Coll (Colledge), Gstn (Gustafson), Rla (Lapatto), Stei (Steiner),
On (Organon), Ubeo (Boehm).
Kiss1r
and
Kiss1
coding regions are numbered and non-coding
exons are not shaded. Promoters are
shaded black
. Deleted exons are indicated by a
delta
symbol.
IRES
, internal ribosome entry sequence;
neo
, promoter driven neomycin resistance gene;
Cre
, cre
recombinase gene;
Gfp
, enhanced green fl uorescent protein gene;
LacZ
,
-galactosidase gene;
NLS
, nuclear localization signal sequence;
rtTA
, reverse tetracyclin-responsive transactivator pro-
tein;
LTR
, proviral long terminal repeat
β
Phenotype of
Kiss1r
and
Kiss1
Global Knock-Out Mice
Analysis of the phenotypes of the
Kiss1r
and
Kiss1
mutant mice has shown the
importance of kisspeptin signalling in the initiation and maintenance of the repro-
ductive axis [
26
,
27
]. In general, the phenotypes of all the mutant mice lines are
Search WWH ::

Custom Search