Environmental Engineering Reference
In-Depth Information
abilities but may also have built-in coding systems for rapid and multiplex target
identification. Nanoparticle labeling is expected to be more sensitive, more flexible in
target selection (e.g., adding new genes or single-nucleotide mutations), more diversified
in bar coding, faster in binding kinetics due to the pseudo-homogeneous reaction, less
expensive to produce and less physically interferential in the biological recognition
events due to the nanoscale-size particles (Han et al., 2001).
Fluorescent
emi ssion
Laser
Laser
Fluorescent
emission
si lica
si lica
RuB py-dye
Surface antigen
antibody
Target DNA
RuBpy-dye
Biotin
Avidin
Capture
probe
Target cell
biochip
Figure 13.5
(a) RuBpy-doped silica nanoparticle based DNA labeling scheme using in
(Lian et al., 2004), (b) RuBpy-doped silica nanoparticle labeling based immunological
scheme used in (Zhao et al., 2004).
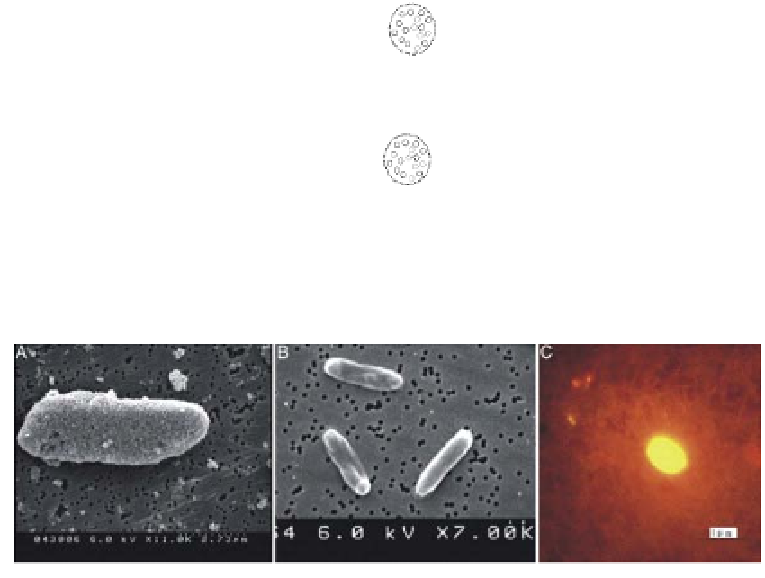
Figure 13.6
(a) Scanning electron microscopy (SEM) image of
E. coli
O157:H7 cell
incubated with dye doped NPs (b) SEM image of
E. coli
DH5 cells incubated with dye
doped NPs (negative control) (c) fluorescence image of a single
E. coli
O157:H7 cell.
Copyright PNAS 2004. Reproduced with permission.
It should be noted, however, that the gain in signal enhancement (hence the
improvement in detection limit) by using nanoparticle-based labeling approaches may
vary by several orders of magnitude at the cell and the DNA level. This is illustrated in
Figure 13.5 and Table 13.3, which compares the enhancement in detection limit from
two studies using similar dye-doped nanoparticle labeling; one uses the label at the cell





































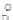







Search WWH ::

Custom Search