Information Technology Reference
In-Depth Information
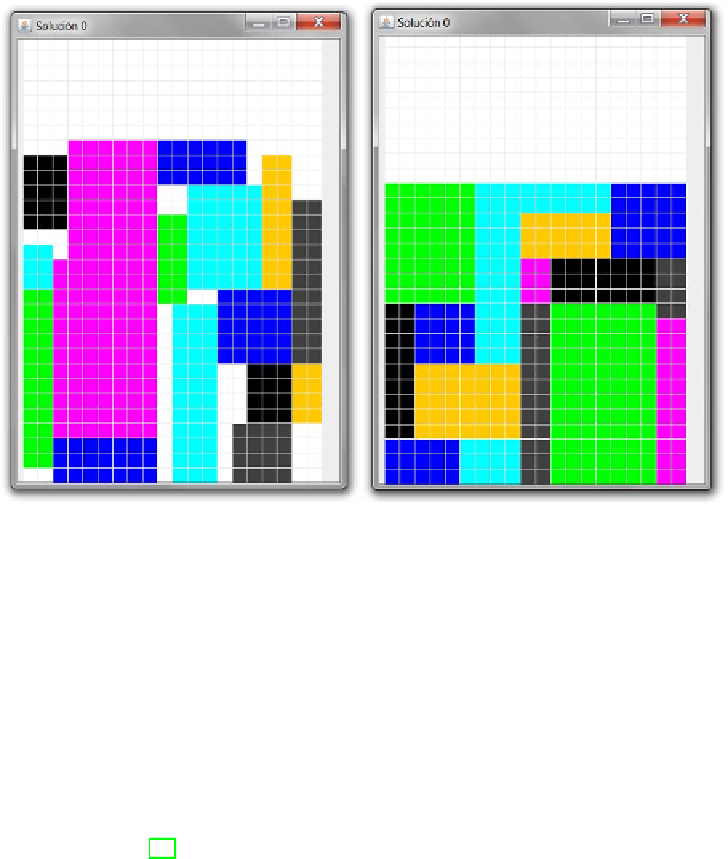
Fig. 2.
Best solution found in the 16-object instance with the automated EA (left),
and the human-guided EA (right)
forcing a highly-greedy decoding process by choosing a short range of values
above zero for genes, or allowing more exploratory strategies by selecting a more
ample value range for genes.
4 Experiments
The experiments have been done with an (
μ, λ
)-EA (
μ
= 100,
λ
= 90,
p
X
=0
.
8,
p
M
=0
.
1). In order to encode the permutation part of the chromosome, a stack-
based encoding [20] is used, i.e., a permutation
p
1
,
···
,p
n
is encoded as an
integer list
e
1
,
i
+ 1); here, a base ordering is
assumed, say,
p
i
=
i
,andthen
e
i
indicates the index of the element of this base
ordering that is selected (without replacement) as the
i
-th element of the permu-
tation. Notice that there is just one choice for the very last element, and hence
the encoding only needs
n
···
,e
n−
1
where 1
e
i
(
n
−
1 values. Furthermore, this encoding is amenable
for simple variation operators, since any sequence represents a valid permuta-
tion as long as each value
e
i
is within the required range. The same applies
to the greedy control sequence, and therefore standard one-point crossover and
random-substitution mutation have been used.
Three problem instances with
n
= 16, 29 and 49 objects have been selected
from [2]. In every case the maximum number of evaluations is set to
maxevals
=
1000
n
. Twenty runs per problem instance are done. In the case of the hgEA,
−
Search WWH ::

Custom Search