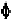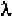Biology Reference
In-Depth Information
CHAPTER 7
STRUCTURES OF POLYNUCLEOTIDES
INTRODUCTION
Similar to polypeptides, periodic structures of polynucleotides can also be
analyzed by steric hindrance studies. The basic building block is a
nucleotide. For RNA, i.e. ribonucleic acid, there are four ribonucleotides
with different bases: adenine (A), cytosine (C), guanine (G) and uracil (U).
For DNA, i.e. deoxyribonucleic acid, there are four deoxyribonucleotides
with different bases: adenine (A), cytosine (C), guanine (G) and thymine
(T). The ribose ring is more or less flat, similar to the peptide bond in
polypeptides, although it can assume several different configurations. The
side chain configurations of nucleotides are specified by one single bond
free to rotate. Unlike the polypeptide backbone where each amino acid
residue has only two single bonds with rotational degree of freedom,
designated by the and angles, polynucleotide backbone has five degrees
of rotational freedom for each nucleotide as shown in Fig. 7-1 (Metzler,
1977). Thus, to define a periodic structure needs at least a five-dimensional
space. For this reason, very few studies on periodic structures of
polynucleotides have been carried out. It is a wide-open field in molecular
biology for detailed theoretical analysis.
In this Chapter, some historical attempts to study polynucleotide structures
are summarized. Nomenclatures used by various authors are different. It is
hoped that once the periodic structures of polynucleotides are understood,
detailed three-dimensional structures of RNA and DNA can be analyzed.
Tremendous amount experimental data are already available for relatively
short RNA and DNA segments. However, even the packing of DNA inside
a -phage is beyond comprehension.